ZFLNCG00024
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592703 | pineal gland | normal | 1007.66 |
| SRR592702 | pineal gland | normal | 831.70 |
| SRR592701 | pineal gland | normal | 755.78 |
| SRR519752 | 7 dpf | VD3 treatment | 345.70 |
| SRR519732 | 7 dpf | FETOH treatment | 318.28 |
| SRR519747 | 6 dpf | VD3 treatment | 318.25 |
| SRR519727 | 6 dpf | FETOH treatment | 287.59 |
| SRR519742 | 4 dpf | VD3 treatment | 266.04 |
| SRR519722 | 4 dpf | FETOH treatment | 253.48 |
| SRR592700 | pineal gland | normal | 237.58 |
Express in tissues
Correlated coding gene
Download
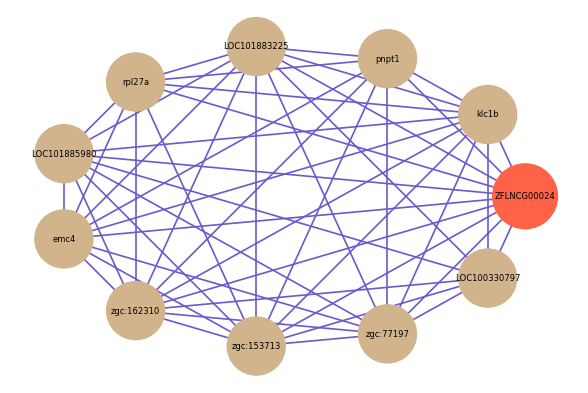
| gene | correlation coefficent |
|---|---|
| klc1b | 0.73 |
| pnpt1 | 0.69 |
| LOC101883225 | 0.66 |
| rpl27a | 0.65 |
| LOC101885980 | 0.65 |
| emc4 | 0.65 |
| zgc:162310 | 0.64 |
| zgc:153713 | 0.64 |
| zgc:77197 | 0.64 |
| LOC100330797 | 0.64 |
Gene Ontology
Download
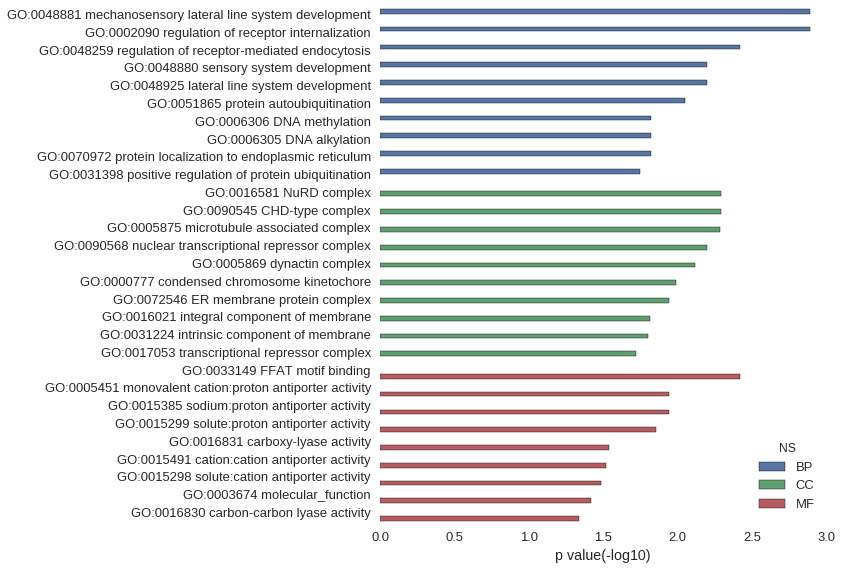
| GO | P value |
|---|---|
| GO:0048881 | 1.28e-03 |
| GO:0002090 | 1.28e-03 |
| GO:0048259 | 3.83e-03 |
| GO:0048880 | 6.38e-03 |
| GO:0048925 | 6.38e-03 |
| GO:0051865 | 8.92e-03 |
| GO:0006306 | 1.52e-02 |
| GO:0006305 | 1.52e-02 |
| GO:0070972 | 1.52e-02 |
| GO:0031398 | 1.78e-02 |
| GO:0016581 | 5.11e-03 |
| GO:0090545 | 5.11e-03 |
| GO:0005875 | 5.18e-03 |
| GO:0090568 | 6.38e-03 |
| GO:0005869 | 7.65e-03 |
| GO:0000777 | 1.02e-02 |
| GO:0072546 | 1.15e-02 |
| GO:0016021 | 1.53e-02 |
| GO:0031224 | 1.58e-02 |
| GO:0017053 | 1.90e-02 |
| GO:0033149 | 3.83e-03 |
| GO:0005451 | 1.15e-02 |
| GO:0015385 | 1.15e-02 |
| GO:0015299 | 1.40e-02 |
| GO:0016831 | 2.90e-02 |
| GO:0015491 | 3.03e-02 |
| GO:0015298 | 3.28e-02 |
| GO:0003674 | 3.84e-02 |
| GO:0016830 | 4.63e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00024
AAAGCACGAACTTTCATCCGTGCTTCATGTTTTGAGGGTAACCCTGTGAATGTGAGTATGTGTGTGTGTT
CATCTGTGTGTGTGTGTTCCTCTATGTGTTCATCTGTGTGTGTGTGTGTGTGTGTACCTGCAGGTCAGAG
AGCATGCCCAGTAGGCTCCTGAGCAGACTGCGGTCCACCGTCTCCCCGCTGCGCTCGCGCTCCACCTGCT
CCAGGATCCCCTGCACCGTCCGGCTCTGAACCGCCGCGTCACTCACTATGTGTGTGCGGAACAGCTCCAG
CCCTGTGTCCCTGTAGCGCACGCGCACACACACACACACACA
AAAGCACGAACTTTCATCCGTGCTTCATGTTTTGAGGGTAACCCTGTGAATGTGAGTATGTGTGTGTGTT
CATCTGTGTGTGTGTGTTCCTCTATGTGTTCATCTGTGTGTGTGTGTGTGTGTGTACCTGCAGGTCAGAG
AGCATGCCCAGTAGGCTCCTGAGCAGACTGCGGTCCACCGTCTCCCCGCTGCGCTCGCGCTCCACCTGCT
CCAGGATCCCCTGCACCGTCCGGCTCTGAACCGCCGCGTCACTCACTATGTGTGTGCGGAACAGCTCCAG
CCCTGTGTCCCTGTAGCGCACGCGCACACACACACACACACA

