ZFLNCG00134
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 3.11 |
| SRR748488 | sperm | normal | 2.44 |
| SRR527835 | tail | normal | 0.79 |
| SRR800037 | egg | normal | 0.76 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 0.74 |
| SRR800046 | sphere stage | 5azaCyD treatment | 0.62 |
| SRR800043 | sphere stage | normal | 0.56 |
| SRR372791 | dome | normal | 0.45 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 0.39 |
| ERR023143 | swim bladder | normal | 0.38 |
Express in tissues
Correlated coding gene
Download
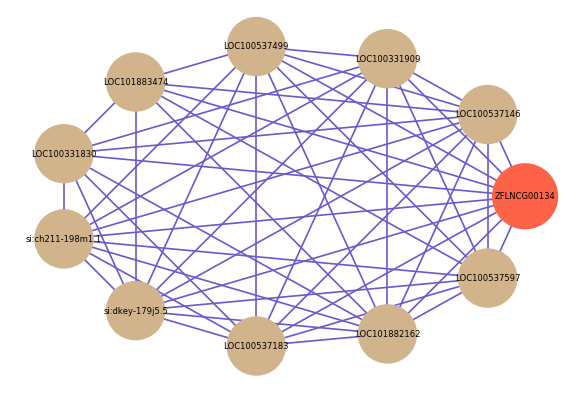
| gene | correlation coefficent |
|---|---|
| LOC100537146 | 0.64 |
| LOC100331909 | 0.61 |
| LOC100537499 | 0.61 |
| LOC101883474 | 0.61 |
| LOC100331830 | 0.60 |
| si:ch211-198m1.1 | 0.60 |
| si:dkey-179j5.5 | 0.60 |
| LOC100537183 | 0.59 |
| LOC101882162 | 0.58 |
| LOC100537597 | 0.57 |
Gene Ontology
Download
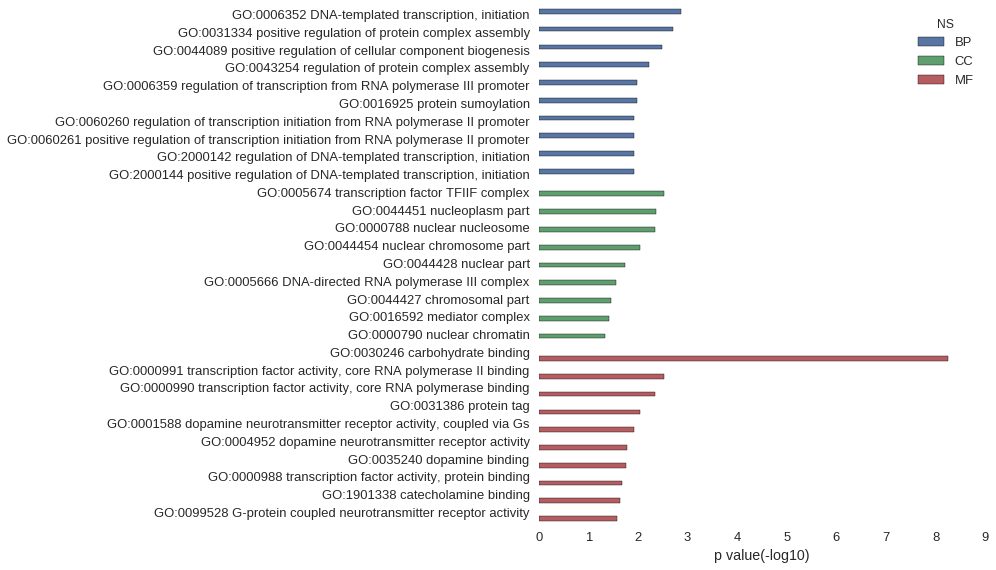
| GO | P value |
|---|---|
| GO:0006352 | 1.37e-03 |
| GO:0031334 | 1.93e-03 |
| GO:0044089 | 3.22e-03 |
| GO:0043254 | 5.95e-03 |
| GO:0006359 | 1.04e-02 |
| GO:0016925 | 1.04e-02 |
| GO:0060260 | 1.19e-02 |
| GO:0060261 | 1.19e-02 |
| GO:2000142 | 1.19e-02 |
| GO:2000144 | 1.19e-02 |
| GO:0005674 | 2.98e-03 |
| GO:0044451 | 4.39e-03 |
| GO:0000788 | 4.47e-03 |
| GO:0044454 | 9.25e-03 |
| GO:0044428 | 1.87e-02 |
| GO:0005666 | 2.80e-02 |
| GO:0044427 | 3.54e-02 |
| GO:0016592 | 3.81e-02 |
| GO:0000790 | 4.53e-02 |
| GO:0030246 | 5.51e-09 |
| GO:0000991 | 2.98e-03 |
| GO:0000990 | 4.47e-03 |
| GO:0031386 | 8.92e-03 |
| GO:0001588 | 1.19e-02 |
| GO:0004952 | 1.63e-02 |
| GO:0035240 | 1.78e-02 |
| GO:0000988 | 2.13e-02 |
| GO:1901338 | 2.36e-02 |
| GO:0099528 | 2.65e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00134
ACCCAAAATTCAAATCTGTATTATCAAACTTTATGCAGCACACTGCATCAAACACTCGACACTAAGCATC
AAACGGCAACTGATCGCCAAATGGATGTCAAGGCACGACTGTCATCAACAAATAATGTTTACTGCTAAAA
ATCTCTTTAGTTTGACTTGCCTAATTGCAGGCAAATTCAGAGAAGTAATAGCATCCGCTCTCAAACGTGT
GAGTCAGTGATGTTTGGACAGAGCGTTTGGAAATTTATGTCCCTGGGTATTGACTGGTTGTTCTTCAAGC
CGCCTGCAGGCATAGATGTGATTCACAATACTCGTTTTGCCCTTATTAGCTGAGGACCTAAACACAAGAG
AAACATTACAGAGTCCCGCTGTGATCTTACACTCATCACCATTAAATAATCATTAAATACACACCTCTTC
TCAGCTCAATCCATCATCAAAGGAAAATTTCCCTAATCTTGTCAGAGGACTCTGGGCTCTGCTCTCTCCT
CAAAATCTACATCTTTCACATTGTAAAGTCTTGAAAACGCTGAAGAGGAAATCAGTAAACAGTCAGGATT
TTGTATTTTTTTAGCAGCTGTTTTACTCTAGTGTGTGTGTGTGTGTGTGTGTGTGTCTGTCGGTCTGTTA
ATTGGTATGTCCT
ACCCAAAATTCAAATCTGTATTATCAAACTTTATGCAGCACACTGCATCAAACACTCGACACTAAGCATC
AAACGGCAACTGATCGCCAAATGGATGTCAAGGCACGACTGTCATCAACAAATAATGTTTACTGCTAAAA
ATCTCTTTAGTTTGACTTGCCTAATTGCAGGCAAATTCAGAGAAGTAATAGCATCCGCTCTCAAACGTGT
GAGTCAGTGATGTTTGGACAGAGCGTTTGGAAATTTATGTCCCTGGGTATTGACTGGTTGTTCTTCAAGC
CGCCTGCAGGCATAGATGTGATTCACAATACTCGTTTTGCCCTTATTAGCTGAGGACCTAAACACAAGAG
AAACATTACAGAGTCCCGCTGTGATCTTACACTCATCACCATTAAATAATCATTAAATACACACCTCTTC
TCAGCTCAATCCATCATCAAAGGAAAATTTCCCTAATCTTGTCAGAGGACTCTGGGCTCTGCTCTCTCCT
CAAAATCTACATCTTTCACATTGTAAAGTCTTGAAAACGCTGAAGAGGAAATCAGTAAACAGTCAGGATT
TTGTATTTTTTTAGCAGCTGTTTTACTCTAGTGTGTGTGTGTGTGTGTGTGTGTGTCTGTCGGTCTGTTA
ATTGGTATGTCCT