ZFLNCG00174
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 50.67 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 44.17 |
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 40.72 |
| ERR023145 | heart | normal | 31.91 |
| SRR1562531 | muscle | normal | 20.28 |
| ERR145648 | skeletal muscle | 27 degree_C to 27 degree_C | 19.98 |
| SRR891510 | muscle | normal | 18.83 |
| SRR372802 | 5 dpf | normal | 14.57 |
| SRR1342219 | embryo | marco morpholino and infection with Mycobacterium marinum | 14.42 |
| ERR023146 | head kidney | normal | 13.30 |
Express in tissues
Correlated coding gene
Download
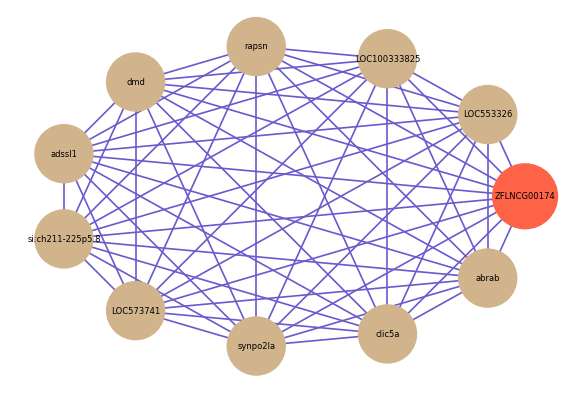
| gene | correlation coefficent |
|---|---|
| LOC553326 | 0.68 |
| LOC100333825 | 0.66 |
| rapsn | 0.64 |
| dmd | 0.62 |
| adssl1 | 0.62 |
| si:ch211-225p5.8 | 0.62 |
| LOC573741 | 0.62 |
| synpo2la | 0.61 |
| clic5a | 0.61 |
| abrab | 0.61 |
Gene Ontology
Download
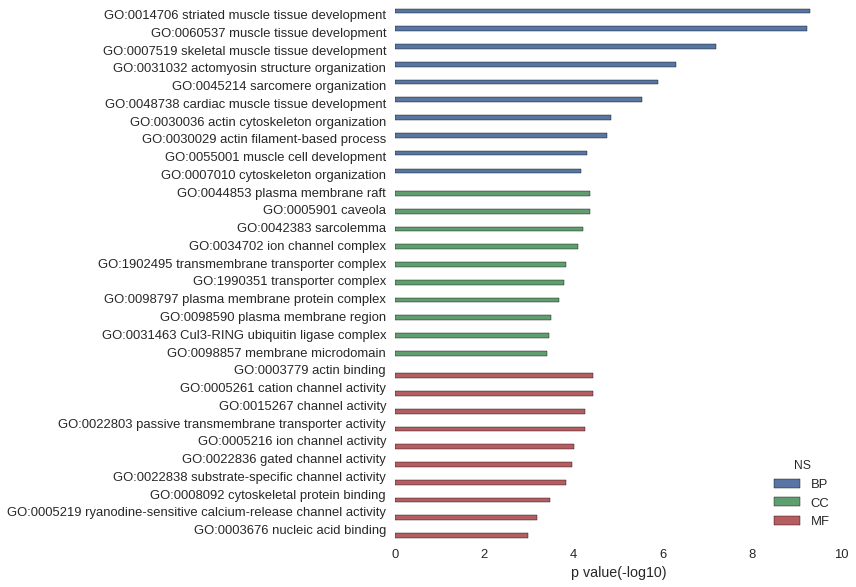
| GO | P value |
|---|---|
| GO:0014706 | 5.00e-10 |
| GO:0060537 | 5.94e-10 |
| GO:0007519 | 6.42e-08 |
| GO:0031032 | 5.16e-07 |
| GO:0045214 | 1.31e-06 |
| GO:0048738 | 2.96e-06 |
| GO:0030036 | 1.47e-05 |
| GO:0030029 | 1.82e-05 |
| GO:0055001 | 5.04e-05 |
| GO:0007010 | 6.91e-05 |
| GO:0044853 | 4.42e-05 |
| GO:0005901 | 4.42e-05 |
| GO:0042383 | 6.28e-05 |
| GO:0034702 | 8.11e-05 |
| GO:1902495 | 1.50e-04 |
| GO:1990351 | 1.64e-04 |
| GO:0098797 | 2.16e-04 |
| GO:0098590 | 3.30e-04 |
| GO:0031463 | 3.51e-04 |
| GO:0098857 | 4.07e-04 |
| GO:0003779 | 3.61e-05 |
| GO:0005261 | 3.75e-05 |
| GO:0015267 | 5.61e-05 |
| GO:0022803 | 5.61e-05 |
| GO:0005216 | 9.96e-05 |
| GO:0022836 | 1.12e-04 |
| GO:0022838 | 1.49e-04 |
| GO:0008092 | 3.44e-04 |
| GO:0005219 | 6.63e-04 |
| GO:0003676 | 1.08e-03 |
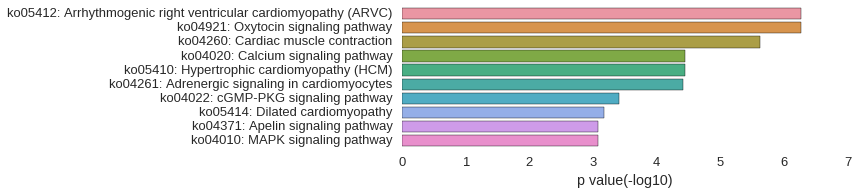
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00174
CAAACACTGACAATTACTTTCCTGAATGTAGTTTACAGACTTAACTTACATTTAATACTTTTACACAGAC
TTTTATGACTTGTGTAAGTATTTTATTTTATCAGTCAGTTCATTCAGTGGTGGAGTCTGTCTGATTTAAT
TGTGTAGCACTTTTGATATTTTTTCCTCCCCAAGCGTGCAATATTATTATTTGAGAACAAATCTCTTTGG
AATTTTATATCTATGTATACAGTCAAGCCTGCAATTATACATACCCTGTA
CAAACACTGACAATTACTTTCCTGAATGTAGTTTACAGACTTAACTTACATTTAATACTTTTACACAGAC
TTTTATGACTTGTGTAAGTATTTTATTTTATCAGTCAGTTCATTCAGTGGTGGAGTCTGTCTGATTTAAT
TGTGTAGCACTTTTGATATTTTTTCCTCCCCAAGCGTGCAATATTATTATTTGAGAACAAATCTCTTTGG
AATTTTATATCTATGTATACAGTCAAGCCTGCAATTATACATACCCTGTA