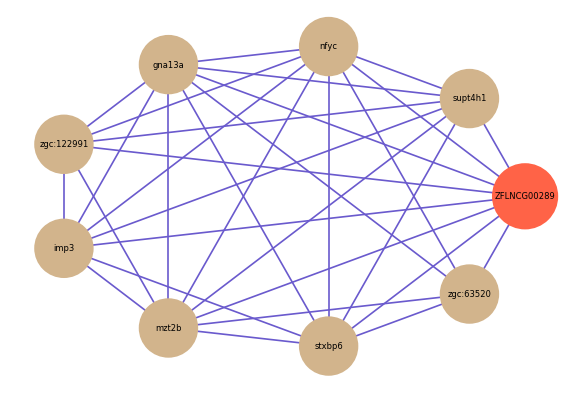
ZFLNCG00289
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR748490 | 1 kcell | normal | 55.11 |
| SRR748488 | sperm | normal | 21.32 |
| SRR1621206 | embryo | normal | 20.51 |
| SRR592701 | pineal gland | normal | 20.36 |
| SRR800046 | sphere stage | 5azaCyD treatment | 18.33 |
| SRR1049814 | 256 cell | normal | 14.45 |
| SRR527835 | tail | normal | 14.08 |
| SRR1021213 | sphere stage | normal | 12.65 |
| SRR800043 | sphere stage | normal | 10.68 |
| SRR1048059 | pineal gland | light | 10.37 |
Express in tissues
Correlated coding gene
Download
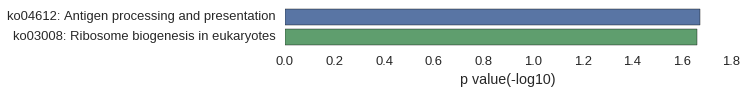
Gene Ontology
Download
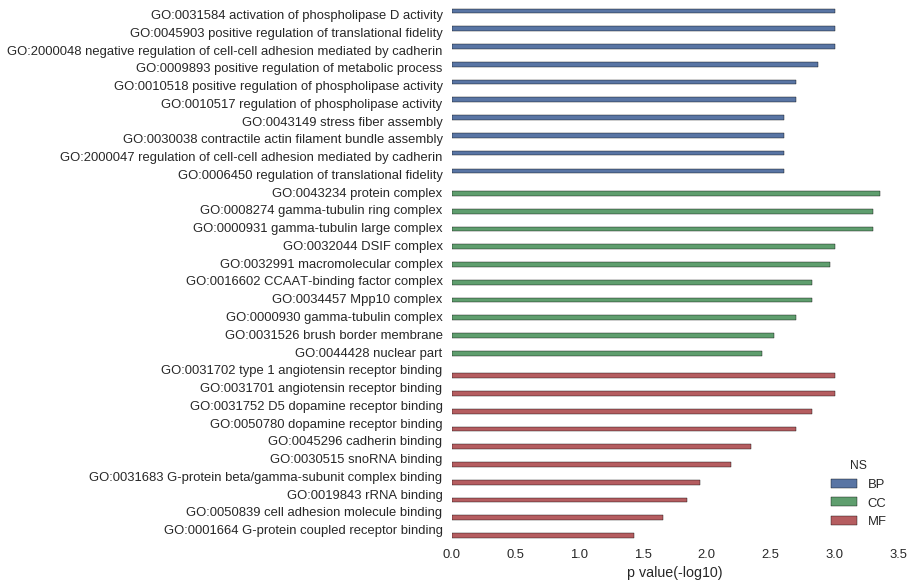
| GO | P value |
|---|---|
| GO:0031584 | 9.95e-04 |
| GO:0045903 | 9.95e-04 |
| GO:2000048 | 9.95e-04 |
| GO:0009893 | 1.35e-03 |
| GO:0010518 | 1.99e-03 |
| GO:0010517 | 1.99e-03 |
| GO:0043149 | 2.49e-03 |
| GO:0030038 | 2.49e-03 |
| GO:2000047 | 2.49e-03 |
| GO:0006450 | 2.49e-03 |
| GO:0043234 | 4.41e-04 |
| GO:0008274 | 4.97e-04 |
| GO:0000931 | 4.97e-04 |
| GO:0032044 | 9.95e-04 |
| GO:0032991 | 1.09e-03 |
| GO:0016602 | 1.49e-03 |
| GO:0034457 | 1.49e-03 |
| GO:0000930 | 1.99e-03 |
| GO:0031526 | 2.98e-03 |
| GO:0044428 | 3.72e-03 |
| GO:0031702 | 9.95e-04 |
| GO:0031701 | 9.95e-04 |
| GO:0031752 | 1.49e-03 |
| GO:0050780 | 1.99e-03 |
| GO:0045296 | 4.47e-03 |
| GO:0030515 | 6.45e-03 |
| GO:0031683 | 1.14e-02 |
| GO:0019843 | 1.43e-02 |
| GO:0050839 | 2.22e-02 |
| GO:0001664 | 3.72e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00289
CTGGAAACTGGTGGAAAGGCGAAGGGGGAAAGCTCCTATGTGCAAACGAACGGTTAGTTTCCTCAGATAT
CCAATCCAGAATTCGGTTAGGATAAACCGGTCTCCCCTCTTCTCCTCTCCGGCTTGCCCGAACGCAAGAT
GAAACGATCCTCTGAAAACGAGCTGAAGTTTTACAAGCACAAACCAAACATTTTCATCCGCCGAAATAAA
GTCTGAGCGTCATTCGCACCCTTCGGAGCCCTGCATCAGGGAGAGCTGAAATTTCTTCGTTATCTTCTGC
AAAAACTCAGATCCCCCATAGAATAATATGTGACCGATCTGAGAAAGCAAGGGATGCGGCACGCGACTCC
GATGCTGCTCCGTTAAAGCGCGATCGCCTTTATCCAACCGATTCCTGTACGACTCCCTGTCCACGGAT
CTGGAAACTGGTGGAAAGGCGAAGGGGGAAAGCTCCTATGTGCAAACGAACGGTTAGTTTCCTCAGATAT
CCAATCCAGAATTCGGTTAGGATAAACCGGTCTCCCCTCTTCTCCTCTCCGGCTTGCCCGAACGCAAGAT
GAAACGATCCTCTGAAAACGAGCTGAAGTTTTACAAGCACAAACCAAACATTTTCATCCGCCGAAATAAA
GTCTGAGCGTCATTCGCACCCTTCGGAGCCCTGCATCAGGGAGAGCTGAAATTTCTTCGTTATCTTCTGC
AAAAACTCAGATCCCCCATAGAATAATATGTGACCGATCTGAGAAAGCAAGGGATGCGGCACGCGACTCC
GATGCTGCTCCGTTAAAGCGCGATCGCCTTTATCCAACCGATTCCTGTACGACTCCCTGTCCACGGAT