ZFLNCG00384
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516123 | skin | male and 3.5 year | 24.80 |
| SRR941749 | anterior pectoral fin | normal | 24.21 |
| SRR516121 | skin | male and 3.5 year | 20.69 |
| SRR516124 | skin | male and 3.5 year | 15.46 |
| SRR1299124 | caudal fin | Zero day time after treatment | 15.02 |
| SRR516131 | skin | male and 5 month | 14.77 |
| SRR658537 | bud | Gata6 morphant | 14.11 |
| SRR527834 | head | normal | 12.01 |
| SRR658539 | bud | Gata5/6 morphant | 11.60 |
| SRR658535 | bud | Gata5 morphant | 11.49 |
Express in tissues
Correlated coding gene
Download
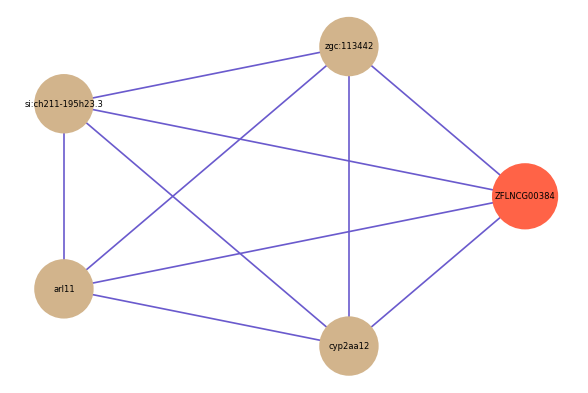
| gene | correlation coefficent |
|---|---|
| zgc:113442 | 0.55 |
| si:ch211-195h23.3 | 0.55 |
| arl11 | 0.54 |
| cyp2aa12 | 0.54 |
Gene Ontology
Download
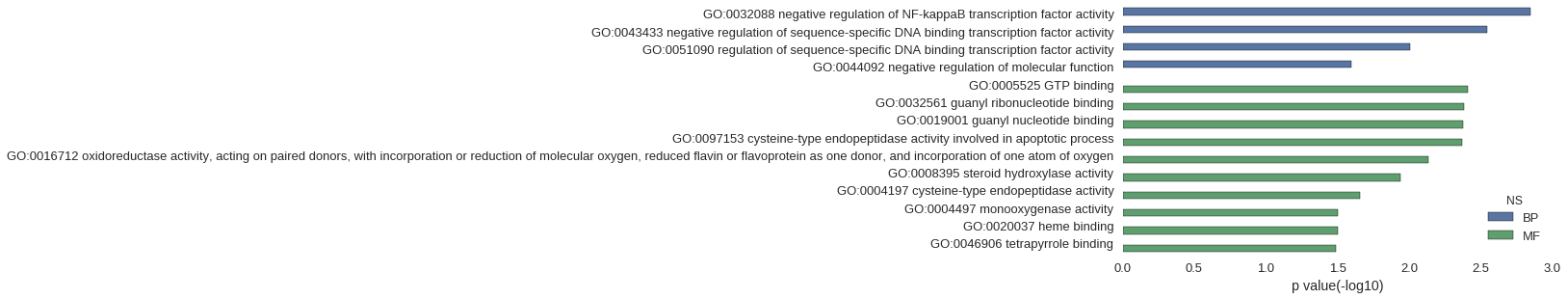
| GO | P value |
|---|---|
| GO:0032088 | 1.42e-03 |
| GO:0043433 | 2.84e-03 |
| GO:0051090 | 9.91e-03 |
| GO:0044092 | 2.53e-02 |
| GO:0005525 | 3.89e-03 |
| GO:0032561 | 4.17e-03 |
| GO:0019001 | 4.19e-03 |
| GO:0097153 | 4.26e-03 |
| GO:0016712 | 7.37e-03 |
| GO:0008395 | 1.16e-02 |
| GO:0004197 | 2.20e-02 |
| GO:0004497 | 3.15e-02 |
| GO:0020037 | 3.15e-02 |
| GO:0046906 | 3.26e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00384
CCCGCCCCTGGAGAATTGTCAGTCTACAATAATCGATTATTGGCTCCTTTACTAGAAGGCAGGACTTCCT
GCACTGCAGTGGCCAAATTGACTGTTACCTTTTTCCCCATTCAAAACTATAGAAGTGACACTTCTTGCGT
GTTCTATGGTCTTTATCCTTACCCAATTTGTCAAACCCAAAAACGCTAGTCTTTCCGAAGTGTTCCAGAT
ACTGTCAATTGTTTTACACTATTAATTTTTGCCTTGGGACATCCATTTTTGTTAACAAATCACCACAACT
TTAACCAGACCATGCCAACATCAGGCTGATATTGTGTAGTCTGAACCCAGCATTAGTGCTTTGCAGATGT
ACAAGAAAAATTCCTGTGTTAGTTCCATTTCTTGTTCCA
CCCGCCCCTGGAGAATTGTCAGTCTACAATAATCGATTATTGGCTCCTTTACTAGAAGGCAGGACTTCCT
GCACTGCAGTGGCCAAATTGACTGTTACCTTTTTCCCCATTCAAAACTATAGAAGTGACACTTCTTGCGT
GTTCTATGGTCTTTATCCTTACCCAATTTGTCAAACCCAAAAACGCTAGTCTTTCCGAAGTGTTCCAGAT
ACTGTCAATTGTTTTACACTATTAATTTTTGCCTTGGGACATCCATTTTTGTTAACAAATCACCACAACT
TTAACCAGACCATGCCAACATCAGGCTGATATTGTGTAGTCTGAACCCAGCATTAGTGCTTTGCAGATGT
ACAAGAAAAATTCCTGTGTTAGTTCCATTTCTTGTTCCA
