ZFLNCG00406
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800037 | egg | normal | 10.20 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 7.13 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 5.71 |
| SRR527835 | tail | normal | 5.33 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 5.16 |
| SRR1021213 | sphere stage | normal | 4.01 |
| SRR1021215 | sphere stage | Mzeomesa | 2.58 |
| SRR748491 | germ ring | normal | 2.56 |
| SRR1621206 | embryo | normal | 2.52 |
| SRR372793 | shield | normal | 2.40 |
Express in tissues
Correlated coding gene
Download
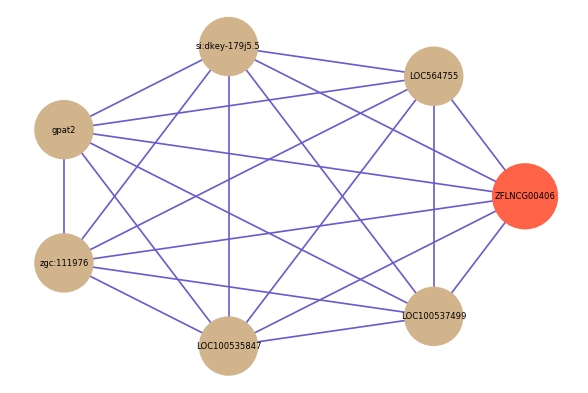
| gene | correlation coefficent |
|---|---|
| LOC564755 | 0.53 |
| si:dkey-179j5.5 | 0.52 |
| gpat2 | 0.51 |
| zgc:111976 | 0.51 |
| LOC100535847 | 0.50 |
| LOC100537499 | 0.50 |
Gene Ontology
Download
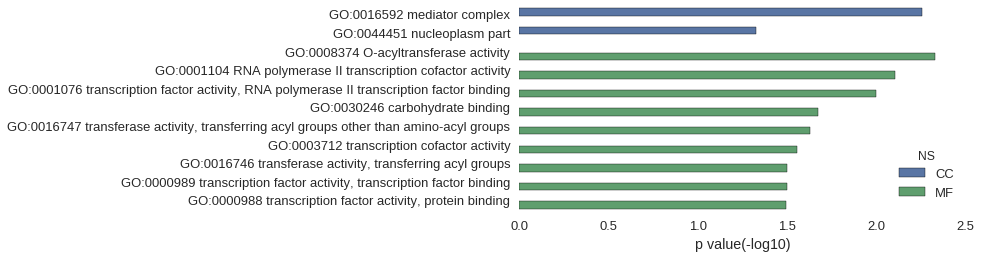
| GO | P value |
|---|---|
| GO:0016592 | 5.53e-03 |
| GO:0044451 | 4.74e-02 |
| GO:0008374 | 4.68e-03 |
| GO:0001104 | 7.87e-03 |
| GO:0001076 | 9.99e-03 |
| GO:0030246 | 2.12e-02 |
| GO:0016747 | 2.35e-02 |
| GO:0003712 | 2.79e-02 |
| GO:0016746 | 3.16e-02 |
| GO:0000989 | 3.16e-02 |
| GO:0000988 | 3.21e-02 |
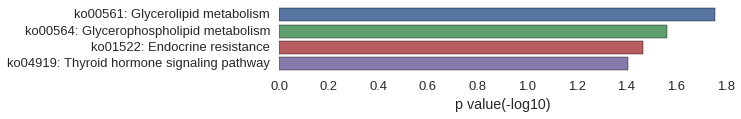
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00406
AAGAGTGTAACTATCATTATGGAAAAGTCATAGAGAATAGAGGTAAGAATGTTTTCAAAATTGATTTGAG
TTTTTATTTCCATCAATATCTATTATTTACTCTTTCTTCTGCCATAGTGCCAGGCGGTGTTGAGACGCGC
TACAGTTGTTGTGAGAATGCAGTTGGGTCACCTGGATGTCAAGTCTTCAATGTATTATAATCTCTGTTCC
AATTATACTCATGCTTTATTAGAGGGACTGAAAACATTTCACCTGATTTGTCTTCACTCTATTATGCTAT
GATTTTTGTCCTTGAACTTTGATTCAATTGTGATATTGTAGCATTTCACCAAAGCCATTAACCTGATGTT
TTCAATGACCAAAGTTGACTTTCAGTGTATTTTCTTGTGTACTTATTTACAGCTTCACGTGCATGATGCA
GTTAGCCTTCAGGGCTTTGTGAGCAGTCTTCATCAGTCTGCGGTGGGAAAAACATGTCCAGGAATCTATG
CCATTGACACACAAACAGTAAGGTTTAAGTACTGATCATAGTTGAGAGTTAAAAAATAATAATAACCAAG
GAAAAAAAGCAGGTAATTTGATATTTAATTTTGTTTTTCACCAGTGTTACACGACTCAAGGTCTGGAACT
AGCAAGAGTGACTGTAGTGAGCAGCAGTTTGCAAGTTGTCTTTGACTCCTTTGTCAAGCCTGATAATGAT
GTCATTGACTATAATACCAGGTAGGGATTGCTTTCAAGGCATAACACTAAATTTGGCAACATTTGAATTG
GATCATCACCTTTCATCAAAGTTTTCCACTGTTTCCTGTTTTAAGACAATTTTGACAAACTTTTTTGACA
TACTTTAAATGTTGATTGGTTTATTTTGCTTTTCACAATGTTGTTTTAGATGAGTGGGGGCATGGTGACA
TT
AAGAGTGTAACTATCATTATGGAAAAGTCATAGAGAATAGAGGTAAGAATGTTTTCAAAATTGATTTGAG
TTTTTATTTCCATCAATATCTATTATTTACTCTTTCTTCTGCCATAGTGCCAGGCGGTGTTGAGACGCGC
TACAGTTGTTGTGAGAATGCAGTTGGGTCACCTGGATGTCAAGTCTTCAATGTATTATAATCTCTGTTCC
AATTATACTCATGCTTTATTAGAGGGACTGAAAACATTTCACCTGATTTGTCTTCACTCTATTATGCTAT
GATTTTTGTCCTTGAACTTTGATTCAATTGTGATATTGTAGCATTTCACCAAAGCCATTAACCTGATGTT
TTCAATGACCAAAGTTGACTTTCAGTGTATTTTCTTGTGTACTTATTTACAGCTTCACGTGCATGATGCA
GTTAGCCTTCAGGGCTTTGTGAGCAGTCTTCATCAGTCTGCGGTGGGAAAAACATGTCCAGGAATCTATG
CCATTGACACACAAACAGTAAGGTTTAAGTACTGATCATAGTTGAGAGTTAAAAAATAATAATAACCAAG
GAAAAAAAGCAGGTAATTTGATATTTAATTTTGTTTTTCACCAGTGTTACACGACTCAAGGTCTGGAACT
AGCAAGAGTGACTGTAGTGAGCAGCAGTTTGCAAGTTGTCTTTGACTCCTTTGTCAAGCCTGATAATGAT
GTCATTGACTATAATACCAGGTAGGGATTGCTTTCAAGGCATAACACTAAATTTGGCAACATTTGAATTG
GATCATCACCTTTCATCAAAGTTTTCCACTGTTTCCTGTTTTAAGACAATTTTGACAAACTTTTTTGACA
TACTTTAAATGTTGATTGGTTTATTTTGCTTTTCACAATGTTGTTTTAGATGAGTGGGGGCATGGTGACA
TT