ZFLNCG00411
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647683 | spleen | SVCV treatment | 40.88 |
| SRR700534 | heart | control morpholino | 37.04 |
| SRR1647684 | spleen | SVCV treatment | 36.37 |
| SRR1291417 | 5 dpi | injected marinum | 24.26 |
| SRR1299125 | caudal fin | Half day time after treatment | 16.19 |
| SRR1299129 | caudal fin | Seven days time after treatment | 15.76 |
| SRR700537 | heart | Gata4 morpholino | 15.30 |
| SRR527834 | head | normal | 14.19 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 14.19 |
| SRR1562529 | intestine and pancreas | normal | 13.51 |
Express in tissues
Correlated coding gene
Download
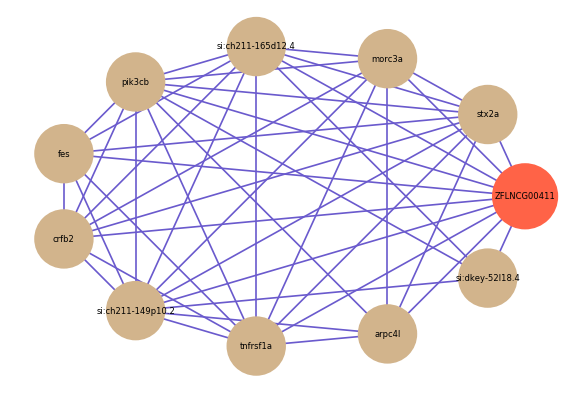
| gene | correlation coefficent |
|---|---|
| stx2a | 0.58 |
| si:dkey-52l18.4 | 0.56 |
| morc3a | 0.56 |
| si:ch211-165d12.4 | 0.56 |
| pik3cb | 0.56 |
| fes | 0.56 |
| crfb2 | 0.56 |
| si:ch211-149p10.2 | 0.55 |
| tnfrsf1a | 0.55 |
| arpc4l | 0.55 |
Gene Ontology
Download
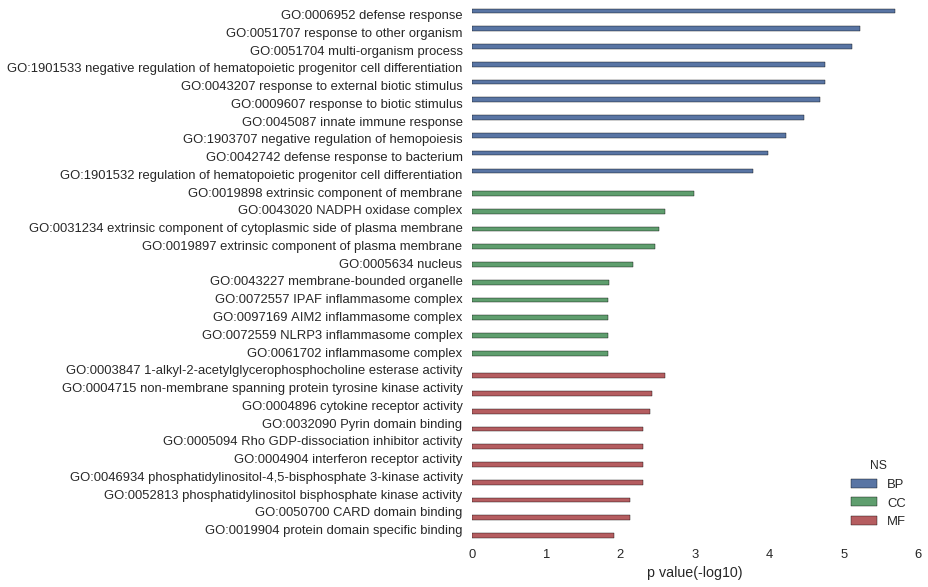
| GO | P value |
|---|---|
| GO:0006952 | 2.02e-06 |
| GO:0051707 | 5.98e-06 |
| GO:0051704 | 7.83e-06 |
| GO:1901533 | 1.80e-05 |
| GO:0043207 | 1.81e-05 |
| GO:0009607 | 2.09e-05 |
| GO:0045087 | 3.44e-05 |
| GO:1903707 | 5.98e-05 |
| GO:0042742 | 1.03e-04 |
| GO:1901532 | 1.67e-04 |
| GO:0019898 | 1.02e-03 |
| GO:0043020 | 2.49e-03 |
| GO:0031234 | 3.02e-03 |
| GO:0019897 | 3.40e-03 |
| GO:0005634 | 6.85e-03 |
| GO:0043227 | 1.42e-02 |
| GO:0072557 | 1.48e-02 |
| GO:0097169 | 1.48e-02 |
| GO:0072559 | 1.48e-02 |
| GO:0061702 | 1.48e-02 |
| GO:0003847 | 2.49e-03 |
| GO:0004715 | 3.79e-03 |
| GO:0004896 | 3.99e-03 |
| GO:0032090 | 4.97e-03 |
| GO:0005094 | 4.97e-03 |
| GO:0004904 | 4.97e-03 |
| GO:0046934 | 4.97e-03 |
| GO:0052813 | 7.44e-03 |
| GO:0050700 | 7.44e-03 |
| GO:0019904 | 1.24e-02 |
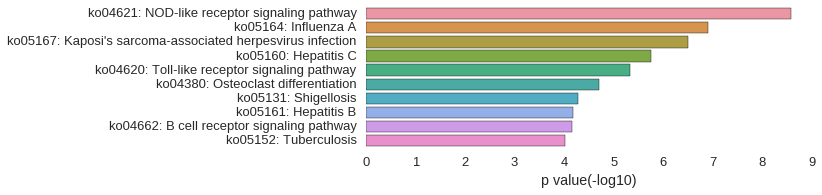
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00411
CTCTCAACATCTTAAAGTCTTCCTTTGGCATCACTGTTATTCCTCAATTTTGTTTTTCGATAAATTCTTT
TAAATTTCTGCAGAAGTGTTCAAACGTGCTGTAGTAGAAGGCCATTTTGTCAGACGCAGCATCTGAGATG
TGAACATTTGATGTCTCAGTTTACCAGGATGTAGGGAACTTCCTCTCTTACTTAAAACTAGAATGTTTTC
AACTACAGAGGGGGGATCAAATGGCAAGTACCTCACAGTCAAAAAGGTGCCTTAATGTCTGAGAAAGTAA
TA
CTCTCAACATCTTAAAGTCTTCCTTTGGCATCACTGTTATTCCTCAATTTTGTTTTTCGATAAATTCTTT
TAAATTTCTGCAGAAGTGTTCAAACGTGCTGTAGTAGAAGGCCATTTTGTCAGACGCAGCATCTGAGATG
TGAACATTTGATGTCTCAGTTTACCAGGATGTAGGGAACTTCCTCTCTTACTTAAAACTAGAATGTTTTC
AACTACAGAGGGGGGATCAAATGGCAAGTACCTCACAGTCAAAAAGGTGCCTTAATGTCTGAGAAAGTAA
TA