ZFLNCG00412
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035237 | liver | transgenic UHRF1 | 2.07 |
| SRR1523214 | embryo | glut12 morpholino | 1.21 |
| SRR1028002 | head | normal | 1.19 |
| SRR1534349 | larvae | DMSO vehicle | 1.00 |
| SRR1534350 | larvae | 5 ug/l BDE47 treatment | 0.87 |
| SRR1534351 | larvae | 500 ug/l BDE47 treatment | 0.77 |
| SRR1291414 | 5 dpi | normal | 0.63 |
| SRR1188160 | embryo | Insulin 4 hpi | 0.34 |
| SRR942769 | embryo | myd88+/+ | 0.33 |
| SRR527834 | head | normal | 0.33 |
Express in tissues
Gene Ontology
Download
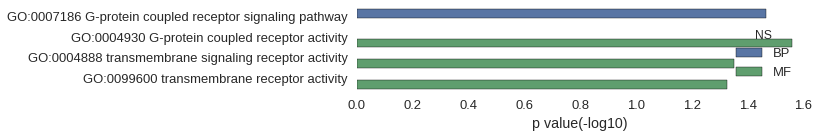
| GO | P value |
|---|---|
| GO:0007186 | 3.41e-02 |
| GO:0004930 | 2.76e-02 |
| GO:0004888 | 4.45e-02 |
| GO:0099600 | 4.70e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT00607 | NONMMUT019050; | Direct blastn |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00412
TGGACTGAAGAGTCAACACTGTAAACTGGACACACTGAGGTCTGAGTTCAAACACCTGATTGATTGATTT
TTTTTTTTTTTGTTTTGTTTTGTTACATCAGACAGATCACATTAACAGTGTGTTTTTAGTAACGTGTGTT
CTTCTTTCTCAGATTGATGACGTGTAAACTCACTGCCGATTCCTGTGAGAGTTTGTCTTCAGCTCTACAA
TCCTCAAACTGTGTGCTGAGAGAGCTGGACCTGAGTATTAATCATCTGCCGGATGCAGGAGTGAAACTTC
TCTCTGATGGACTGAAGAGTCAACACTGTACACTGGAGACACTGAGGTCTGAATTTAAACACCTGATTGA
TTGATTGATTGATTGAATAAATTTTTGTTTTGTTACATCAGGCAGATTACATTAACAGTGTGGTTTTTAG
TAATCTGTGTTCTTCTTTCTCAGATTAGTCATGTGTAATCTCACTGTTCAGTGCTG
TGGACTGAAGAGTCAACACTGTAAACTGGACACACTGAGGTCTGAGTTCAAACACCTGATTGATTGATTT
TTTTTTTTTTTGTTTTGTTTTGTTACATCAGACAGATCACATTAACAGTGTGTTTTTAGTAACGTGTGTT
CTTCTTTCTCAGATTGATGACGTGTAAACTCACTGCCGATTCCTGTGAGAGTTTGTCTTCAGCTCTACAA
TCCTCAAACTGTGTGCTGAGAGAGCTGGACCTGAGTATTAATCATCTGCCGGATGCAGGAGTGAAACTTC
TCTCTGATGGACTGAAGAGTCAACACTGTACACTGGAGACACTGAGGTCTGAATTTAAACACCTGATTGA
TTGATTGATTGATTGAATAAATTTTTGTTTTGTTACATCAGGCAGATTACATTAACAGTGTGGTTTTTAG
TAATCTGTGTTCTTCTTTCTCAGATTAGTCATGTGTAATCTCACTGTTCAGTGCTG