ZFLNCG00457
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516133 | skin | male and 5 month | 128.01 |
| SRR516125 | skin | male and 3.5 year | 87.19 |
| SRR527834 | head | normal | 48.58 |
| SRR726542 | 5 dpf | infection with control | 41.63 |
| SRR726540 | 5 dpf | normal | 36.52 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 35.43 |
| SRR516123 | skin | male and 3.5 year | 33.70 |
| SRR519727 | 6 dpf | FETOH treatment | 28.11 |
| SRR527833 | 5 dpf | normal | 23.23 |
| SRR800045 | muscle | normal | 21.89 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| LOC101882980 | 0.58 |
| fam46bb | 0.58 |
| LOC100005636 | 0.58 |
| LOC100000002 | 0.58 |
| si:dkey-30j22.2 | 0.58 |
| LOC100330638 | 0.58 |
| LOC568975 | 0.57 |
| LOC560023 | 0.57 |
| LOC101883885 | 0.56 |
| LOC568930 | 0.56 |
Gene Ontology
Download
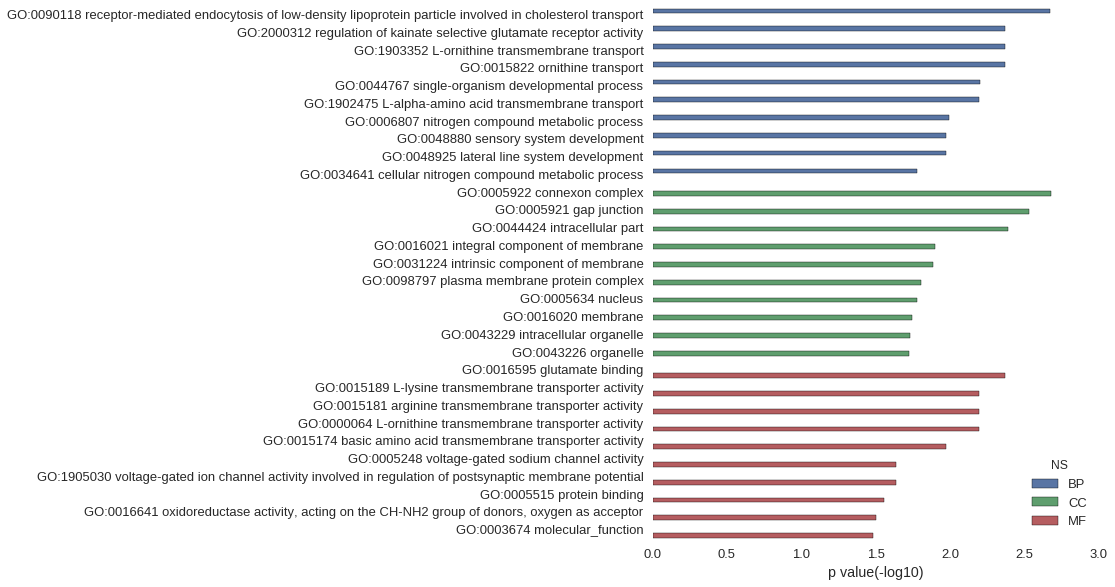
| GO | P value |
|---|---|
| GO:0090118 | 2.13e-03 |
| GO:2000312 | 4.26e-03 |
| GO:1903352 | 4.26e-03 |
| GO:0015822 | 4.26e-03 |
| GO:0044767 | 6.29e-03 |
| GO:1902475 | 6.38e-03 |
| GO:0006807 | 1.02e-02 |
| GO:0048880 | 1.06e-02 |
| GO:0048925 | 1.06e-02 |
| GO:0034641 | 1.66e-02 |
| GO:0005922 | 2.09e-03 |
| GO:0005921 | 2.94e-03 |
| GO:0044424 | 4.08e-03 |
| GO:0016021 | 1.27e-02 |
| GO:0031224 | 1.30e-02 |
| GO:0098797 | 1.56e-02 |
| GO:0005634 | 1.67e-02 |
| GO:0016020 | 1.80e-02 |
| GO:0043229 | 1.86e-02 |
| GO:0043226 | 1.89e-02 |
| GO:0016595 | 4.26e-03 |
| GO:0015189 | 6.38e-03 |
| GO:0015181 | 6.38e-03 |
| GO:0000064 | 6.38e-03 |
| GO:0015174 | 1.06e-02 |
| GO:0005248 | 2.32e-02 |
| GO:1905030 | 2.32e-02 |
| GO:0005515 | 2.77e-02 |
| GO:0016641 | 3.15e-02 |
| GO:0003674 | 3.30e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00457
AGTATTTCTACATTTGTTGTTGGTGTTGTTTCCACAGCTGAAGACACTGTTGAGGTGGTTGTGGCAGCAT
ACGTTTTCGTTATCGATTCCTCAGTAGTTGTTGTGGCTGGAGAGGTTGTTGTGGTTGTTGTATCAGGTGA
TGTAGTCATAGTATCGGGATTAGTTGTTGATGAGGATTCGACAGTTGATGATAGTGAAATGGTGGATGTC
ACATCAGAAGTTGATGTTGTTGCCCCACCAACTGTTGTGGTTTCAGCTGTAGTTGTTCTTGGTGTTTCAG
GTAATGTAGTTAATATTTCTACATTTGTTGTTGGTGTTGTTTCCACAGTTGAAGACACTGTTGAGGTGGT
TGTGGCAACATACATTTTTGTTGTCGTTTCCTCAGTAGTTGTTGTAGCTGAAGAGGTTGTTGTAGTTGTT
GTATTAGGTGATATAGTCATAGTGTCTGCATTAGTCGTTGATTCCACAATTGAGGATACTGTTGATGTGG
ATGTGGCATTA
AGTATTTCTACATTTGTTGTTGGTGTTGTTTCCACAGCTGAAGACACTGTTGAGGTGGTTGTGGCAGCAT
ACGTTTTCGTTATCGATTCCTCAGTAGTTGTTGTGGCTGGAGAGGTTGTTGTGGTTGTTGTATCAGGTGA
TGTAGTCATAGTATCGGGATTAGTTGTTGATGAGGATTCGACAGTTGATGATAGTGAAATGGTGGATGTC
ACATCAGAAGTTGATGTTGTTGCCCCACCAACTGTTGTGGTTTCAGCTGTAGTTGTTCTTGGTGTTTCAG
GTAATGTAGTTAATATTTCTACATTTGTTGTTGGTGTTGTTTCCACAGTTGAAGACACTGTTGAGGTGGT
TGTGGCAACATACATTTTTGTTGTCGTTTCCTCAGTAGTTGTTGTAGCTGAAGAGGTTGTTGTAGTTGTT
GTATTAGGTGATATAGTCATAGTGTCTGCATTAGTCGTTGATTCCACAATTGAGGATACTGTTGATGTGG
ATGTGGCATTA

