ZFLNCG00494
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 126.28 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 122.89 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 106.95 |
| SRR1647682 | spleen | normal | 67.35 |
| SRR1048059 | pineal gland | light | 67.31 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 64.98 |
| SRR1647683 | spleen | SVCV treatment | 63.75 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 57.12 |
| SRR372802 | 5 dpf | normal | 43.47 |
| SRR1205166 | 5dpf | transgenic sqET20 and neomycin treated 3h | 41.23 |
Express in tissues
Correlated coding gene
Download
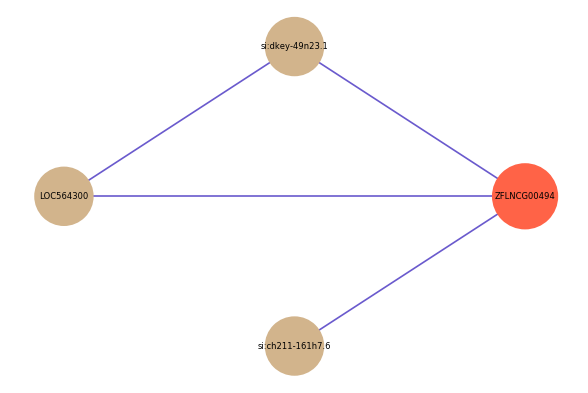
| gene | correlation coefficent |
|---|---|
| si:dkey-49n23.1 | 0.56 |
| si:ch211-161h7.6 | 0.52 |
| LOC564300 | 0.51 |
Gene Ontology
Download
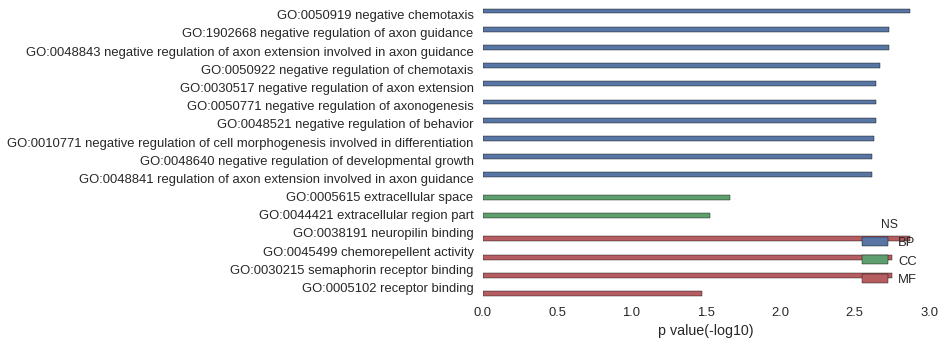
| GO | P value |
|---|---|
| GO:0050919 | 1.35e-03 |
| GO:1902668 | 1.85e-03 |
| GO:0048843 | 1.85e-03 |
| GO:0050922 | 2.13e-03 |
| GO:0030517 | 2.27e-03 |
| GO:0050771 | 2.27e-03 |
| GO:0048521 | 2.27e-03 |
| GO:0010771 | 2.35e-03 |
| GO:0048640 | 2.42e-03 |
| GO:0048841 | 2.42e-03 |
| GO:0005615 | 2.17e-02 |
| GO:0044421 | 2.95e-02 |
| GO:0038191 | 1.35e-03 |
| GO:0045499 | 1.78e-03 |
| GO:0030215 | 1.78e-03 |
| GO:0005102 | 3.38e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00494
TTTCAGGACATCAGTTGCCTTTCTGCTGACTTGAAAATGCCCAGCACACAAGCTCATTCTGCTGACACGG
ATCCTCCCAAATTCCACCGTCTCTTTAGGTTCCATGACCCTGAAGTGGTGGCAGTAAGTAAAGACGATAA
CAGATAAAGAAAACAGTAATAGATAAATAAATTATACAGTATTTAACTGTAACATGAACTGTATTTTACA
GTAAGACAGTTTTGCAGTGTGTTACTGTATTTTAAAGTTGCAATAAGAAAATGACTGTATATTTTACAGT
AATAATATACAATACTGTAAATACAAATAGAGCATATAGGCCTCGTGTTTGATTGGGGTTGGAGCAAAAC
TACAGAGCTGCGGCCCTCCAGGAATTGAGTTTGAGACCTATGATATCGAGCAAGATAAATGGTGCTGTAA
ATGTAAATACAGTATGTGTGCTGTAATTGATTTACAGTAAGTTACTACAGAGTCTACAGGAAATGAACAG
TGTAGAGATGTTTTGCTTAATGTTGCTGTTGTTTAATGTGTGCATTTTTCTAGGTTATGGCAATACTGCT
AGGTCTGTTCCAGCTGCTCCTGACTTTATTAACTCACACCGTCAGCCTTGACATGAAGTATATGTTCGTG
TGCCCCCTTTGTGTTGGTTCTGT
TTTCAGGACATCAGTTGCCTTTCTGCTGACTTGAAAATGCCCAGCACACAAGCTCATTCTGCTGACACGG
ATCCTCCCAAATTCCACCGTCTCTTTAGGTTCCATGACCCTGAAGTGGTGGCAGTAAGTAAAGACGATAA
CAGATAAAGAAAACAGTAATAGATAAATAAATTATACAGTATTTAACTGTAACATGAACTGTATTTTACA
GTAAGACAGTTTTGCAGTGTGTTACTGTATTTTAAAGTTGCAATAAGAAAATGACTGTATATTTTACAGT
AATAATATACAATACTGTAAATACAAATAGAGCATATAGGCCTCGTGTTTGATTGGGGTTGGAGCAAAAC
TACAGAGCTGCGGCCCTCCAGGAATTGAGTTTGAGACCTATGATATCGAGCAAGATAAATGGTGCTGTAA
ATGTAAATACAGTATGTGTGCTGTAATTGATTTACAGTAAGTTACTACAGAGTCTACAGGAAATGAACAG
TGTAGAGATGTTTTGCTTAATGTTGCTGTTGTTTAATGTGTGCATTTTTCTAGGTTATGGCAATACTGCT
AGGTCTGTTCCAGCTGCTCCTGACTTTATTAACTCACACCGTCAGCCTTGACATGAAGTATATGTTCGTG
TGCCCCCTTTGTGTTGGTTCTGT
