ZFLNCG00496
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516124 | skin | male and 3.5 year | 275.28 |
| SRR1299126 | caudal fin | One day time after treatment | 205.39 |
| SRR372802 | 5 dpf | normal | 175.37 |
| SRR516125 | skin | male and 3.5 year | 161.36 |
| SRR516123 | skin | male and 3.5 year | 152.13 |
| SRR592702 | pineal gland | normal | 148.37 |
| SRR1534351 | larvae | 500 ug/l BDE47 treatment | 119.24 |
| SRR1534350 | larvae | 5 ug/l BDE47 treatment | 118.05 |
| SRR941753 | posterior pectoral fin | normal | 108.23 |
| SRR941749 | anterior pectoral fin | normal | 105.26 |
Express in tissues
Correlated coding gene
Download
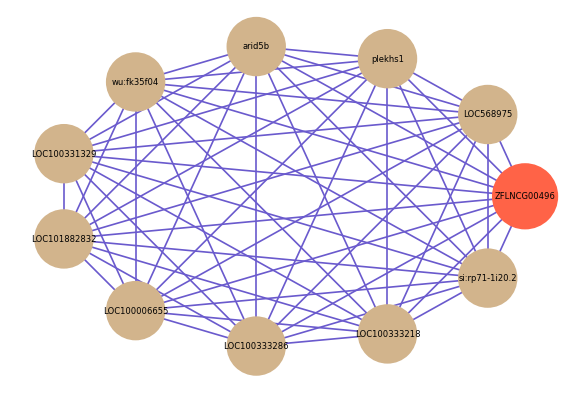
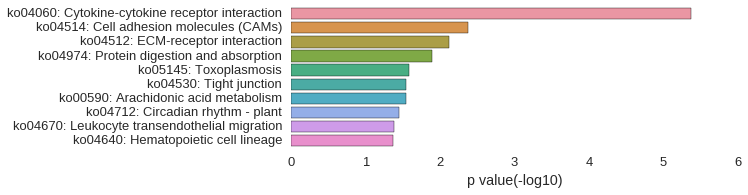
| gene | correlation coefficent |
|---|---|
| LOC568975 | 0.67 |
| plekhs1 | 0.66 |
| arid5b | 0.66 |
| wu:fk35f04 | 0.66 |
| LOC100331329 | 0.65 |
| LOC101882832 | 0.65 |
| LOC100006655 | 0.64 |
| LOC100333286 | 0.64 |
| LOC100333218 | 0.64 |
| si:rp71-1i20.2 | 0.64 |
Gene Ontology
Download
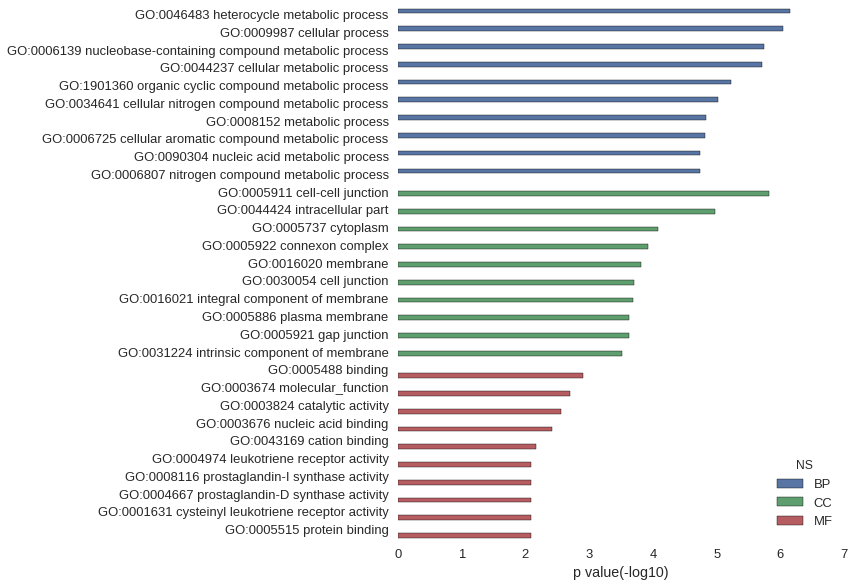
| GO | P value |
|---|---|
| GO:0046483 | 7.01e-07 |
| GO:0009987 | 9.18e-07 |
| GO:0006139 | 1.84e-06 |
| GO:0044237 | 1.96e-06 |
| GO:1901360 | 6.05e-06 |
| GO:0034641 | 9.47e-06 |
| GO:0008152 | 1.49e-05 |
| GO:0006725 | 1.50e-05 |
| GO:0090304 | 1.82e-05 |
| GO:0006807 | 1.82e-05 |
| GO:0005911 | 1.53e-06 |
| GO:0044424 | 1.05e-05 |
| GO:0005737 | 8.41e-05 |
| GO:0005922 | 1.19e-04 |
| GO:0016020 | 1.56e-04 |
| GO:0030054 | 2.00e-04 |
| GO:0016021 | 2.04e-04 |
| GO:0005886 | 2.33e-04 |
| GO:0005921 | 2.36e-04 |
| GO:0031224 | 3.05e-04 |
| GO:0005488 | 1.23e-03 |
| GO:0003674 | 1.98e-03 |
| GO:0003824 | 2.76e-03 |
| GO:0003676 | 3.83e-03 |
| GO:0043169 | 6.92e-03 |
| GO:0004974 | 8.03e-03 |
| GO:0008116 | 8.03e-03 |
| GO:0004667 | 8.03e-03 |
| GO:0001631 | 8.03e-03 |
| GO:0005515 | 8.12e-03 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00496
CGGGCTCTAAATGCGAAAGAAAGACTAGTAAATGATAAAGCTAGATCATATTTTCACTTGTTTACACCTT
GATTTTCTGATTCTGAGTGTGACCACAGCCATGATTATTGTGTAGTAGCATGATGTCGTGTTTTTGCTGA
AGTGAATGTGAAGCTTAGTGCATCACAGCTGAAAACAGAAGGGATGCTAAGGTCATCCACAATGCTGTTA
GCATACTAGCCAGCACCGGAGACTTGTTGG
CGGGCTCTAAATGCGAAAGAAAGACTAGTAAATGATAAAGCTAGATCATATTTTCACTTGTTTACACCTT
GATTTTCTGATTCTGAGTGTGACCACAGCCATGATTATTGTGTAGTAGCATGATGTCGTGTTTTTGCTGA
AGTGAATGTGAAGCTTAGTGCATCACAGCTGAAAACAGAAGGGATGCTAAGGTCATCCACAATGCTGTTA
GCATACTAGCCAGCACCGGAGACTTGTTGG