ZFLNCG00574
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR594771 | endothelium | normal | 2.38 |
| ERR023144 | brain | normal | 1.18 |
| SRR1021215 | sphere stage | Mzeomesa | 0.78 |
| SRR372802 | 5 dpf | normal | 0.64 |
| SRR535890 | larvae | normal | 0.60 |
| SRR1021213 | sphere stage | normal | 0.57 |
| SRR1562533 | testis | normal | 0.48 |
| SRR592698 | pineal gland | normal | 0.46 |
| SRR658537 | bud | Gata6 morphant | 0.44 |
| SRR535926 | larvae | normal | 0.43 |
Express in tissues
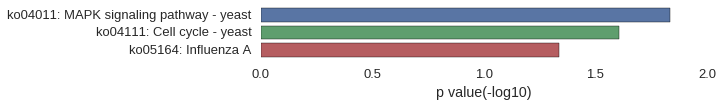
Correlated coding gene
Download
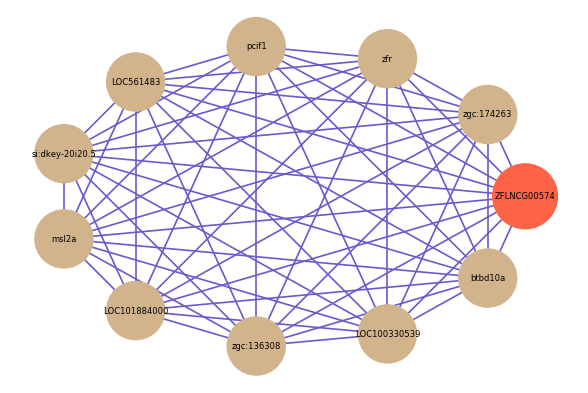
| gene | correlation coefficent |
|---|---|
| zgc:174263 | 0.54 |
| zfr | 0.54 |
| pcif1 | 0.53 |
| LOC561483 | 0.52 |
| si:dkey-20i20.5 | 0.52 |
| msl2a | 0.52 |
| LOC101884000 | 0.52 |
| zgc:136308 | 0.51 |
| LOC100330539 | 0.51 |
| btbd10a | 0.51 |
Gene Ontology
Download
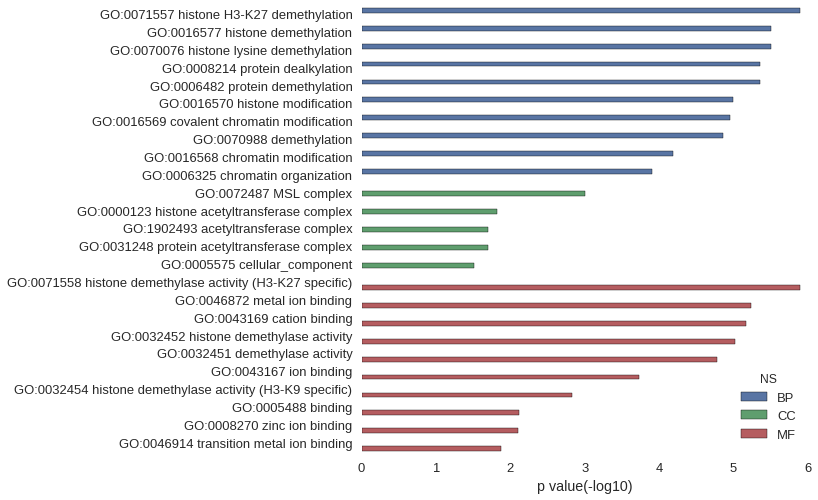
| GO | P value |
|---|---|
| GO:0071557 | 1.27e-06 |
| GO:0016577 | 3.18e-06 |
| GO:0070076 | 3.18e-06 |
| GO:0008214 | 4.45e-06 |
| GO:0006482 | 4.45e-06 |
| GO:0016570 | 1.02e-05 |
| GO:0016569 | 1.12e-05 |
| GO:0070988 | 1.40e-05 |
| GO:0016568 | 6.49e-05 |
| GO:0006325 | 1.24e-04 |
| GO:0072487 | 9.95e-04 |
| GO:0000123 | 1.53e-02 |
| GO:1902493 | 1.97e-02 |
| GO:0031248 | 1.97e-02 |
| GO:0005575 | 3.05e-02 |
| GO:0071558 | 1.27e-06 |
| GO:0046872 | 5.88e-06 |
| GO:0043169 | 6.77e-06 |
| GO:0032452 | 9.53e-06 |
| GO:0032451 | 1.65e-05 |
| GO:0043167 | 1.86e-04 |
| GO:0032454 | 1.49e-03 |
| GO:0005488 | 7.61e-03 |
| GO:0008270 | 7.84e-03 |
| GO:0046914 | 1.33e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00574
TCTGCAGGGTGGAGGAAGAAGTAGTGGAGCCTGCAAAGTAAAAAAAGAAACATGTTATAGCTTTAGTCAA
AACATGTCAAATAAACCATAAACACAACGTCCTTCTGGTTTCCTCACCCTCAGTGAGGTCCTCGATTGCC
TTCCTCACGCCGCGATCAAACGCTCGAGCGTCAGCGGGACTCTGGAACGACAGACCGCACTTCCTGTTGT
CCACTCGCCAGTGATGGAAAGTGGGCGTGGCTTTAGTGTAGACCAGATCTTTCTTCAGAAAGCACTGCAG
GATCATCTGCTCAACATGCACAGACAGCGTTATCGGGATTGATGCTTCATGTTTTTCAACACAGGCTCAT
CCTTAATACGTGATTCTGGCTCTGTTTATGTAGATCCTGAAATGCTTTCCCATGTTTTAATGCACTTTTC
TGTTGCACCATTTTTGTCGGTATGTGAATCCGGCTGTAAACTGAAATGAATTGCTACTGGTATGTTTCAT
TGCATTTTTAGTAATGAAATGTCAACAAGAGGGTTTGGAACCACTTGAGTGAGAGCAAACAGTAGGGTTG
TCCAGATCTGATCATGTGATCATAAATCGCACCCAATCACGTGGATTCAGACTTCGTTAACTCCCAAT
TCTGCAGGGTGGAGGAAGAAGTAGTGGAGCCTGCAAAGTAAAAAAAGAAACATGTTATAGCTTTAGTCAA
AACATGTCAAATAAACCATAAACACAACGTCCTTCTGGTTTCCTCACCCTCAGTGAGGTCCTCGATTGCC
TTCCTCACGCCGCGATCAAACGCTCGAGCGTCAGCGGGACTCTGGAACGACAGACCGCACTTCCTGTTGT
CCACTCGCCAGTGATGGAAAGTGGGCGTGGCTTTAGTGTAGACCAGATCTTTCTTCAGAAAGCACTGCAG
GATCATCTGCTCAACATGCACAGACAGCGTTATCGGGATTGATGCTTCATGTTTTTCAACACAGGCTCAT
CCTTAATACGTGATTCTGGCTCTGTTTATGTAGATCCTGAAATGCTTTCCCATGTTTTAATGCACTTTTC
TGTTGCACCATTTTTGTCGGTATGTGAATCCGGCTGTAAACTGAAATGAATTGCTACTGGTATGTTTCAT
TGCATTTTTAGTAATGAAATGTCAACAAGAGGGTTTGGAACCACTTGAGTGAGAGCAAACAGTAGGGTTG
TCCAGATCTGATCATGTGATCATAAATCGCACCCAATCACGTGGATTCAGACTTCGTTAACTCCCAAT