ZFLNCG00605
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 105.95 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 73.06 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 71.13 |
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 21.24 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 19.05 |
| ERR023143 | swim bladder | normal | 13.51 |
| ERR023144 | brain | normal | 12.89 |
| ERR023146 | head kidney | normal | 9.08 |
| SRR516124 | skin | male and 3.5 year | 7.92 |
| ERR023145 | heart | normal | 6.97 |
Express in tissues
Correlated coding gene
Download
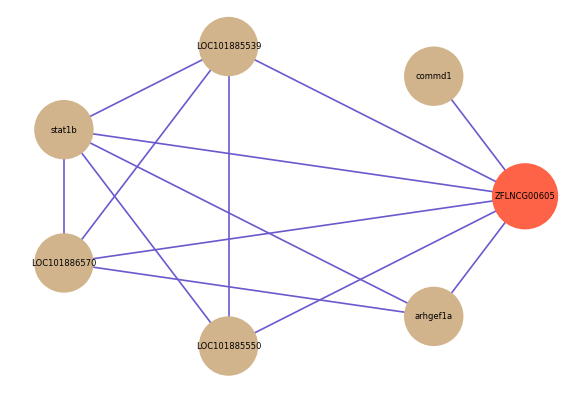
| gene | correlation coefficent |
|---|---|
| commd1 | 0.63 |
| LOC101885539 | 0.53 |
| stat1b | 0.53 |
| LOC101886570 | 0.51 |
| arhgef1a | 0.50 |
| LOC101885550 | 0.50 |
Gene Ontology
Download
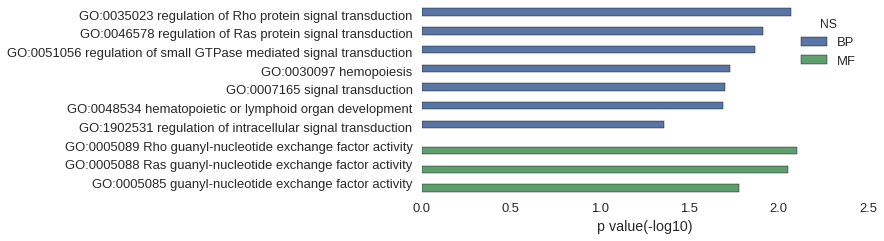
| GO | P value |
|---|---|
| GO:0035023 | 8.51e-03 |
| GO:0046578 | 1.22e-02 |
| GO:0051056 | 1.36e-02 |
| GO:0030097 | 1.88e-02 |
| GO:0007165 | 1.99e-02 |
| GO:0048534 | 2.06e-02 |
| GO:1902531 | 4.37e-02 |
| GO:0005089 | 7.94e-03 |
| GO:0005088 | 8.93e-03 |
| GO:0005085 | 1.67e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00605
ATGACTCAAGATGATGTCTGCGTTATAATTACCATTTATAGCTTGTAGCATACAGGAACAGTCTTATCTA
ATGTATCTTTGACTTGACTTTCAGAGCGATGACCTTTGCTCAAAAGCAACAGCGCTGATAAGACAGATTA
AGATATTATAGTGGATCTCTGCACTTCCTTGAACACTAAATCAGTCGGTGGAGATGATAATCATTAACTC
AAATTAAAGAGACAGTTGACCATTATTTACTCACACTTTACTGGCTCCAAACCAGTTTGAGTTGATTAAT
TTTGCTGTTCAATACAAAAGAAGATACTTTGAAGAATGTTTGAGTGTTCTTAAATGTGCTGCCGAATCAA
GATGGGATTTACTCATTATCACATATAAAGCTTTGCTGGGATCAAAACTCATGACAAAGACTTTGTACAG
GTTCATTCAAGCACATCATCTACAGAGCTGCTGTGTGTGATGTGTCAAGCATTGATAGCAGACACACGCC
AAAATAACACACAACCTGAAATACTGTGATCATTTCTTGAAAAATAATGATTTAAAAAACTAAACCGATA
TCAAAGACTGAGATTTCACCATGATTTTTTACTTGAATAACGTACAGCATCAAACACACACACACATACA
CACACACACACACACACACACACACACACACACACACACACACACACACACACACACACACACACACAAA
CACACACAAACACACACAAACACACACAAACACACACAAACACACACACACACACACACACACACACACA
CACACGCGCATGTGCTTATACACAAACTCTCTCACACACACAGGTAAACATACAAACAAACCCAAACACA
CTTCTACACGCACACACACACACAGGCAGACCATAACACACACACAAAACTCACACACACACATATATGC
TGTGCTTACACCCAAACTCTCTCACACACACACAGGCATGCACACAGGTAAACATACAAACAAACCCAAA
CACACTTCTACACGCACACACACAGGCACACCATCACACACACACACACACACACACACACACACACACA
CACACACACACACACACACACACACACACACACACACATGCTGTGCTTACACACAAACTCTCTCACACAC
ACAGGCATGCACAAAGGTAAACATACAAACAAACCCAAACACACTTCTACACGCACACACACACACACAG
GCACACCATCACACAC
ATGACTCAAGATGATGTCTGCGTTATAATTACCATTTATAGCTTGTAGCATACAGGAACAGTCTTATCTA
ATGTATCTTTGACTTGACTTTCAGAGCGATGACCTTTGCTCAAAAGCAACAGCGCTGATAAGACAGATTA
AGATATTATAGTGGATCTCTGCACTTCCTTGAACACTAAATCAGTCGGTGGAGATGATAATCATTAACTC
AAATTAAAGAGACAGTTGACCATTATTTACTCACACTTTACTGGCTCCAAACCAGTTTGAGTTGATTAAT
TTTGCTGTTCAATACAAAAGAAGATACTTTGAAGAATGTTTGAGTGTTCTTAAATGTGCTGCCGAATCAA
GATGGGATTTACTCATTATCACATATAAAGCTTTGCTGGGATCAAAACTCATGACAAAGACTTTGTACAG
GTTCATTCAAGCACATCATCTACAGAGCTGCTGTGTGTGATGTGTCAAGCATTGATAGCAGACACACGCC
AAAATAACACACAACCTGAAATACTGTGATCATTTCTTGAAAAATAATGATTTAAAAAACTAAACCGATA
TCAAAGACTGAGATTTCACCATGATTTTTTACTTGAATAACGTACAGCATCAAACACACACACACATACA
CACACACACACACACACACACACACACACACACACACACACACACACACACACACACACACACACACAAA
CACACACAAACACACACAAACACACACAAACACACACAAACACACACACACACACACACACACACACACA
CACACGCGCATGTGCTTATACACAAACTCTCTCACACACACAGGTAAACATACAAACAAACCCAAACACA
CTTCTACACGCACACACACACACAGGCAGACCATAACACACACACAAAACTCACACACACACATATATGC
TGTGCTTACACCCAAACTCTCTCACACACACACAGGCATGCACACAGGTAAACATACAAACAAACCCAAA
CACACTTCTACACGCACACACACAGGCACACCATCACACACACACACACACACACACACACACACACACA
CACACACACACACACACACACACACACACACACACACATGCTGTGCTTACACACAAACTCTCTCACACAC
ACAGGCATGCACAAAGGTAAACATACAAACAAACCCAAACACACTTCTACACGCACACACACACACACAG
GCACACCATCACACAC