ZFLNCG00633
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 85.79 |
| SRR1035978 | 13 hpf | rx3-/- | 79.79 |
| SRR941753 | posterior pectoral fin | normal | 75.13 |
| SRR658539 | bud | Gata5/6 morphant | 70.98 |
| SRR658537 | bud | Gata6 morphant | 70.08 |
| SRR658533 | bud | normal | 68.54 |
| SRR891495 | heart | normal | 68.43 |
| SRR594769 | blood | normal | 66.49 |
| SRR891510 | muscle | normal | 66.16 |
| SRR891504 | liver | normal | 62.38 |
Express in tissues
Correlated coding gene
Download
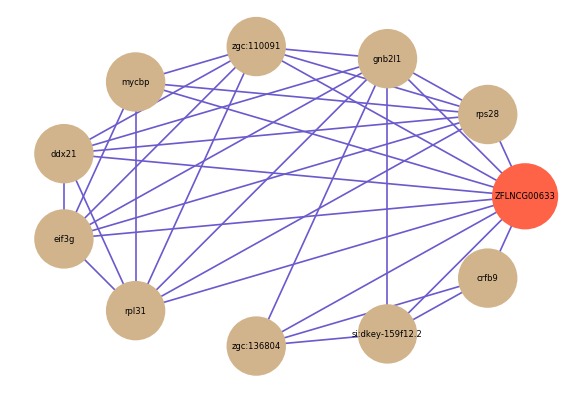
| gene | correlation coefficent |
|---|---|
| rps28 | 0.64 |
| zgc:136804 | 0.62 |
| gnb2l1 | 0.60 |
| zgc:110091 | 0.59 |
| si:dkey-159f12.2 | 0.59 |
| mycbp | 0.58 |
| ddx21 | 0.58 |
| eif3g | 0.58 |
| rpl31 | 0.58 |
| crfb9 | 0.57 |
Gene Ontology
Download
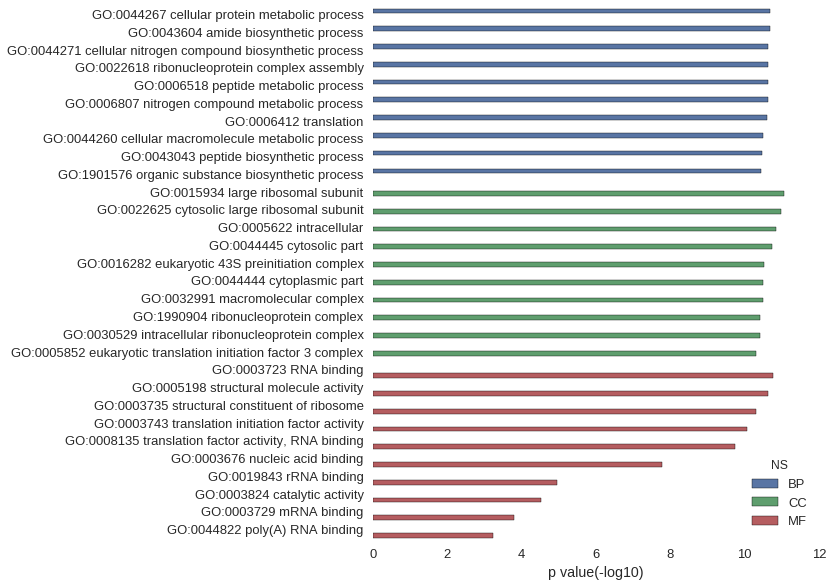
| GO | P value |
|---|---|
| GO:0044267 | 2.03e-11 |
| GO:0043604 | 2.03e-11 |
| GO:0044271 | 2.29e-11 |
| GO:0022618 | 2.29e-11 |
| GO:0006518 | 2.32e-11 |
| GO:0006807 | 2.36e-11 |
| GO:0006412 | 2.49e-11 |
| GO:0044260 | 3.24e-11 |
| GO:0043043 | 3.36e-11 |
| GO:1901576 | 3.53e-11 |
| GO:0015934 | 8.62e-12 |
| GO:0022625 | 1.06e-11 |
| GO:0005622 | 1.44e-11 |
| GO:0044445 | 1.90e-11 |
| GO:0016282 | 2.96e-11 |
| GO:0044444 | 3.24e-11 |
| GO:0032991 | 3.29e-11 |
| GO:1990904 | 3.80e-11 |
| GO:0030529 | 3.80e-11 |
| GO:0005852 | 4.91e-11 |
| GO:0003723 | 1.74e-11 |
| GO:0005198 | 2.39e-11 |
| GO:0003735 | 4.86e-11 |
| GO:0003743 | 8.89e-11 |
| GO:0008135 | 1.84e-10 |
| GO:0003676 | 1.66e-08 |
| GO:0019843 | 1.08e-05 |
| GO:0003824 | 2.94e-05 |
| GO:0003729 | 1.62e-04 |
| GO:0044822 | 5.98e-04 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00633
TTATTTTATCATTTTATTAAGTTAAAATAGCAAAGATTATGGCTAACTGTAGGGGTGTTTTAACTATCCG
TTCAAACAGTGTCGATCGTCAGTCTTTTCCCGCGCGGGGAGCTCCGCCTAGCGGCTGCACGAAAGATGAC
ACGCTCCTGATTACTTTGCTTCATTTAGAGGGTCCAGCTGCAGGTTTAGGGGCTCCTGCTGTCCCGCTTT
TCTGTGCTGGAGTTTTCAACACACTCAGTTTCTTATGCAGGGACGTGCTGGCCTTCTGTGTGACTTCATG
TTCAATCTTGTTCCTGATGGCCACCTCCAAACCCTGCGCGTGCACACACACACCACTATTATTCAACTAG
AAGGGGTTTAAACAGGACTAAACTGGCTATATAGCTTATTTCATACCTTCTTTAGCTTCTGTTGTTGGAC
CACTTGAGCTTTTTTGGGAGCGATTATCCTTCCTGGAATAAAAAGTAAACAGATATTGAGTGCGCATGCG
CATTTACACATGAACAAACACGCTACTTTTTTTACACTGTATGTTTTTAGCGTTTAAAATACTCATATTT
TAAAGTAATACGATGTTTTTACCTC
TTATTTTATCATTTTATTAAGTTAAAATAGCAAAGATTATGGCTAACTGTAGGGGTGTTTTAACTATCCG
TTCAAACAGTGTCGATCGTCAGTCTTTTCCCGCGCGGGGAGCTCCGCCTAGCGGCTGCACGAAAGATGAC
ACGCTCCTGATTACTTTGCTTCATTTAGAGGGTCCAGCTGCAGGTTTAGGGGCTCCTGCTGTCCCGCTTT
TCTGTGCTGGAGTTTTCAACACACTCAGTTTCTTATGCAGGGACGTGCTGGCCTTCTGTGTGACTTCATG
TTCAATCTTGTTCCTGATGGCCACCTCCAAACCCTGCGCGTGCACACACACACCACTATTATTCAACTAG
AAGGGGTTTAAACAGGACTAAACTGGCTATATAGCTTATTTCATACCTTCTTTAGCTTCTGTTGTTGGAC
CACTTGAGCTTTTTTGGGAGCGATTATCCTTCCTGGAATAAAAAGTAAACAGATATTGAGTGCGCATGCG
CATTTACACATGAACAAACACGCTACTTTTTTTACACTGTATGTTTTTAGCGTTTAAAATACTCATATTT
TAAAGTAATACGATGTTTTTACCTC

