ZFLNCG00663
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR891512 | blood | normal | 27.62 |
| SRR592699 | pineal gland | normal | 9.83 |
| SRR516125 | skin | male and 3.5 year | 6.01 |
| SRR726542 | 5 dpf | infection with control | 5.66 |
| ERR023146 | head kidney | normal | 5.57 |
| SRR1299125 | caudal fin | Half day time after treatment | 5.55 |
| SRR1299128 | caudal fin | Three days time after treatment | 5.29 |
| SRR1035984 | 13 hpf | normal | 4.10 |
| SRR516121 | skin | male and 3.5 year | 4.08 |
| SRR372796 | bud | normal | 3.73 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| LOC100148249 | 0.61 |
| rnf183 | 0.59 |
| zgc:110333 | 0.57 |
| si:ch211-95j8.2 | 0.55 |
| LOC558513 | 0.55 |
| krt17 | 0.55 |
| LOC100000832 | 0.54 |
| btr30 | 0.54 |
| ch25hl1.1 | 0.53 |
| si:dkey-17e16.17 | 0.53 |
Gene Ontology
Download
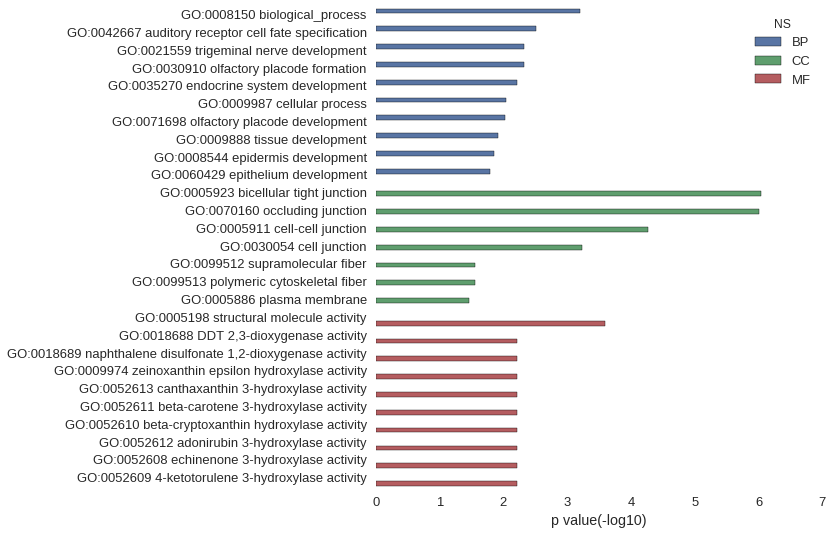
| GO | P value |
|---|---|
| GO:0008150 | 6.40e-04 |
| GO:0042667 | 3.12e-03 |
| GO:0021559 | 4.68e-03 |
| GO:0030910 | 4.68e-03 |
| GO:0035270 | 6.24e-03 |
| GO:0009987 | 8.97e-03 |
| GO:0071698 | 9.35e-03 |
| GO:0009888 | 1.24e-02 |
| GO:0008544 | 1.40e-02 |
| GO:0060429 | 1.63e-02 |
| GO:0005923 | 9.06e-07 |
| GO:0070160 | 9.84e-07 |
| GO:0005911 | 5.49e-05 |
| GO:0030054 | 5.78e-04 |
| GO:0099512 | 2.83e-02 |
| GO:0099513 | 2.83e-02 |
| GO:0005886 | 3.44e-02 |
| GO:0005198 | 2.52e-04 |
| GO:0018688 | 6.24e-03 |
| GO:0018689 | 6.24e-03 |
| GO:0009974 | 6.24e-03 |
| GO:0052613 | 6.24e-03 |
| GO:0052611 | 6.24e-03 |
| GO:0052610 | 6.24e-03 |
| GO:0052612 | 6.24e-03 |
| GO:0052608 | 6.24e-03 |
| GO:0052609 | 6.24e-03 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00663
CTGGGTTTGTTTTACTTGAGCTTTTGACTTCACAGCATTAGTTTTCCTCTTGTTGTATGTTGTGATTGTA
TGTTATGGCGTGTGTAACCACTAGATGGCACCAGAGGTTTGGAAAGGGTTATGTGAGATGAGAGAGTGCT
AGAATAAGACAGTATCAGGCAGACTGAACTTGTAAATATAGAAGACACTCGTCCAACTCGTCGTCATTAT
TATAAGACGCAAACATGAGATGGTACCAGAGCATAGCAAGACAAATGCC
CTGGGTTTGTTTTACTTGAGCTTTTGACTTCACAGCATTAGTTTTCCTCTTGTTGTATGTTGTGATTGTA
TGTTATGGCGTGTGTAACCACTAGATGGCACCAGAGGTTTGGAAAGGGTTATGTGAGATGAGAGAGTGCT
AGAATAAGACAGTATCAGGCAGACTGAACTTGTAAATATAGAAGACACTCGTCCAACTCGTCGTCATTAT
TATAAGACGCAAACATGAGATGGTACCAGAGCATAGCAAGACAAATGCC