ZFLNCG00721
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR372787 | 2-4 cell | normal | 7.42 |
| SRR038627 | embryo | control morpholino | 6.80 |
| SRR658535 | bud | Gata5 morphant | 6.55 |
| SRR658545 | 6 somite | Gata6 morphant | 6.37 |
| SRR658541 | 6 somite | normal | 5.97 |
| SRR1035978 | 13 hpf | rx3-/- | 5.84 |
| SRR658543 | 6 somite | Gata5 morphan | 5.82 |
| SRR658547 | 6 somite | Gata5/6 morphant | 5.79 |
| SRR038625 | embryo | traf6 morpholino | 5.51 |
| SRR658533 | bud | normal | 5.49 |
Express in tissues
Correlated coding gene
Download
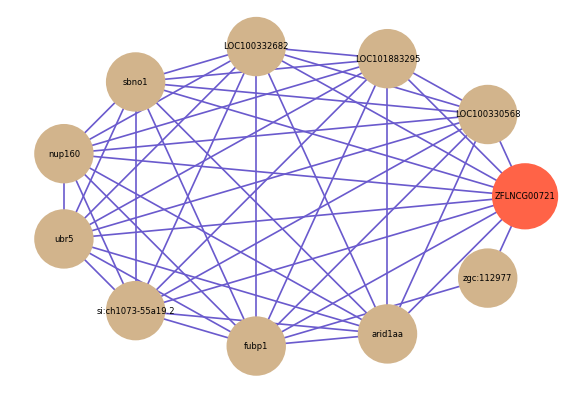
| gene | correlation coefficent |
|---|---|
| LOC100330568 | 0.72 |
| LOC101883295 | 0.70 |
| LOC100332682 | 0.69 |
| sbno1 | 0.69 |
| zgc:112977 | 0.68 |
| nup160 | 0.68 |
| ubr5 | 0.67 |
| si:ch1073-55a19.2 | 0.67 |
| fubp1 | 0.67 |
| arid1aa | 0.65 |
Gene Ontology
Download
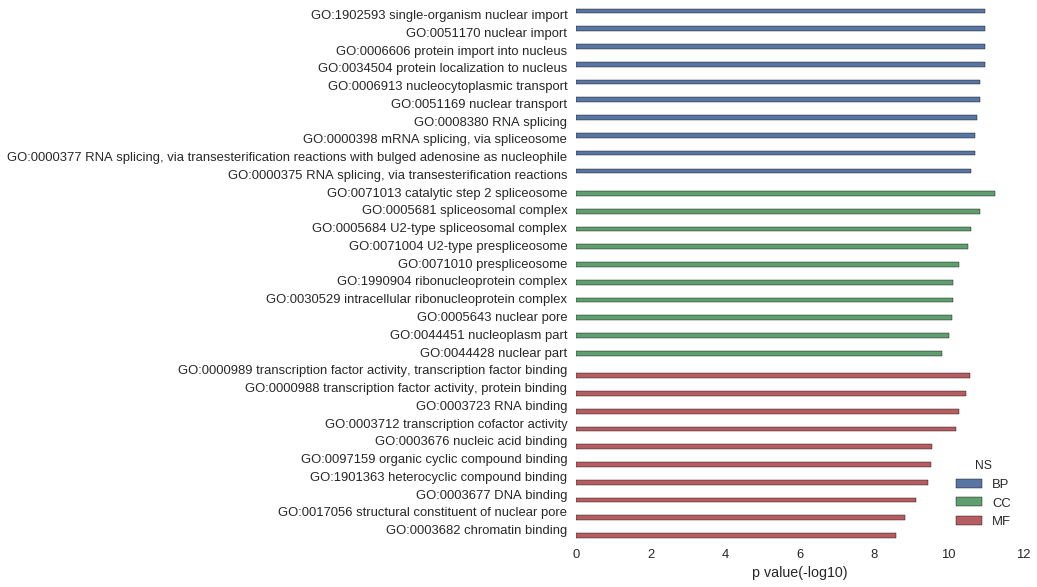
| GO | P value |
|---|---|
| GO:1902593 | 1.03e-11 |
| GO:0051170 | 1.03e-11 |
| GO:0006606 | 1.03e-11 |
| GO:0034504 | 1.03e-11 |
| GO:0006913 | 1.46e-11 |
| GO:0051169 | 1.46e-11 |
| GO:0008380 | 1.73e-11 |
| GO:0000398 | 1.88e-11 |
| GO:0000377 | 1.88e-11 |
| GO:0000375 | 2.47e-11 |
| GO:0071013 | 5.73e-12 |
| GO:0005681 | 1.46e-11 |
| GO:0005684 | 2.54e-11 |
| GO:0071004 | 2.99e-11 |
| GO:0071010 | 5.30e-11 |
| GO:1990904 | 7.75e-11 |
| GO:0030529 | 7.75e-11 |
| GO:0005643 | 8.19e-11 |
| GO:0044451 | 9.72e-11 |
| GO:0044428 | 1.50e-10 |
| GO:0000989 | 2.58e-11 |
| GO:0000988 | 3.47e-11 |
| GO:0003723 | 5.14e-11 |
| GO:0003712 | 6.42e-11 |
| GO:0003676 | 2.73e-10 |
| GO:0097159 | 2.90e-10 |
| GO:1901363 | 3.55e-10 |
| GO:0003677 | 7.45e-10 |
| GO:0017056 | 1.49e-09 |
| GO:0003682 | 2.54e-09 |
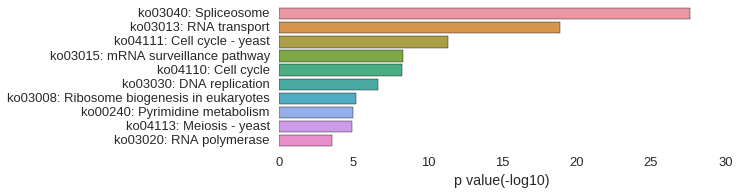
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00721
CTTTCGGTTGAGGAAGTTGCTGGCGACGCGGCACAGCATCAGCAGGCTGTCCTCGGTGTGTGTCCAGGCC
ACTCTCTGTCTGGTCATGTGTAGCAGCGCACGCTGATCTGTCTCATCCTGAGCCTGTCTCTGCCTCTGCA
CGCGCACTGAACACACACACACACACACACACACACACACACACACACACACGCACGCACGCACGCATGC
ACACACGCACACACACACACGCACAGTTGATCTACTGGTCATTGTGTCTGTGTCATGCAGTGTGTGTGTG
TGTGTGTGTGTGTGTGTGTACCTCTCCTCTTCTTGCGTGGTGGTTTCTCCTTCTTCTGCCTCTTGCGCTT
CAGGTTTCGTCCTCCGCTATGCTGCTGGATCCTGCTGGACGGCTCTGAACTTATGCGCACCTGCTCCTCC
AGCACAGCAGGGGGCGACACCTTTACTTCCACTGCGTGCACACACACACACACACACACACACACACACA
CACACCACAGGACTGAGCAATCGGACTCAACGAGGCTGCATCTGAAATCACATCACTGTGTGTGTGCGTG
TGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTTACCTGTGTTTACAGCAGGTTTGGGCAGTGGGTGTTTGG
TCAGCAGGTTCCTCAGTCTGACAGTCTGTTGAGCTCCTGCGTCATCAGTGCTCTG
CTTTCGGTTGAGGAAGTTGCTGGCGACGCGGCACAGCATCAGCAGGCTGTCCTCGGTGTGTGTCCAGGCC
ACTCTCTGTCTGGTCATGTGTAGCAGCGCACGCTGATCTGTCTCATCCTGAGCCTGTCTCTGCCTCTGCA
CGCGCACTGAACACACACACACACACACACACACACACACACACACACACACGCACGCACGCACGCATGC
ACACACGCACACACACACACGCACAGTTGATCTACTGGTCATTGTGTCTGTGTCATGCAGTGTGTGTGTG
TGTGTGTGTGTGTGTGTGTACCTCTCCTCTTCTTGCGTGGTGGTTTCTCCTTCTTCTGCCTCTTGCGCTT
CAGGTTTCGTCCTCCGCTATGCTGCTGGATCCTGCTGGACGGCTCTGAACTTATGCGCACCTGCTCCTCC
AGCACAGCAGGGGGCGACACCTTTACTTCCACTGCGTGCACACACACACACACACACACACACACACACA
CACACCACAGGACTGAGCAATCGGACTCAACGAGGCTGCATCTGAAATCACATCACTGTGTGTGTGCGTG
TGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTTACCTGTGTTTACAGCAGGTTTGGGCAGTGGGTGTTTGG
TCAGCAGGTTCCTCAGTCTGACAGTCTGTTGAGCTCCTGCGTCATCAGTGCTCTG