ZFLNCG00723
Basic Information
Chromesome: chr1
Start: 60288388
End: 60289386
Transcript: ZFLNCT01071 ZFLNCT01072 ZFLNCT01073
Known as:
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR658543 | 6 somite | Gata5 morphan | 15.86 |
| SRR527832 | 16-36 hpf | normal | 8.96 |
| SRR658537 | bud | Gata6 morphant | 8.94 |
| SRR038625 | embryo | traf6 morpholino | 8.58 |
| SRR658533 | bud | normal | 8.35 |
| SRR658539 | bud | Gata5/6 morphant | 7.39 |
| SRR658547 | 6 somite | Gata5/6 morphant | 6.37 |
| SRR658535 | bud | Gata5 morphant | 5.97 |
| SRR749514 | 24 hpf | normal | 5.61 |
| SRR700534 | heart | control morpholino | 5.22 |
Express in tissues
Correlated coding gene
Download
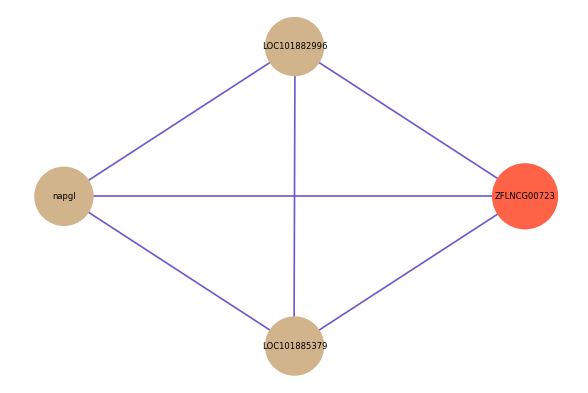
| gene | correlation coefficent |
|---|---|
| LOC101882996 | 0.52 |
| napgl | 0.50 |
| LOC101885379 | 0.50 |
Gene Ontology
Gene Ontology of this lncRNA gene hava does not exist!
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00723
CATGTCAGACACGAGAGAATTTAGACATTGTTTTGGAGAATCTACATTATAGAAATCAATGCTGAGACTC
ACATCTACATCATGTGTTATGATCAGCAGTATATGATATAATATTGATATTCATCAGTGTATTTACCCGC
AGGTGAGTGTTATGGGCAGTAATATATGATATAATATTGATATTCATCAGTGTATTTACCCGCAGGTGTG
TGTTATGATCAGCAGTATATGATATAATATTGATATTCATCAGTGTATTTACCCGCAGGTGAGTGTTATG
GGCAGTAATATATGATATAATATTGATATTCATCAGTGTATTTACCAGCAGGTGAGTGTTATGATCAGCA
GTATATGATATAATATTGATATTCATCAGTGTATTTACCCGCAGGTGAGTGTTATGGGCAGTAATATATG
ATATAATATTGATATTCATCAGTGTATTTACCCGCAGGTGAGTGTTATGGGCAGTAATATATGATATAAT
ATTGATATTCATCAGTGTATTTACCAGCAGGTGAGTGTTATGATCAGCAGTATATGATATAATATTGATA
TTCATCAGTGTATTTACCAGCAGGTGAGTGTTATGGGCAGTAATATATGATATAATATTGATATTCATCA
GTGTATTTACCAGCAGGTGAGTGTTATGATCAGCAGTATATGATATAATATTGATATTCATCAGTGTATT
TACCCGCAGGTGTGTGTTATGATCAGCAGTATATGATATAATATTGATATTCATCAGTGTATTTACCCGC
AGGTGAGTGTTATGGGCAGCAGGAGCGCGCAGGTCAGCAGTACGGCACCGCAGCACAGTATCTTTACGGG
TCTTCTCGCCATCTGATTCACCGCCTGCCGGAGCTCAGCCCGAACCCGCCGCAGCATCACCGACCGCTAC
AGACAGCAGCTCATCCCGGTGAACCGAACACAGCTGAACAGGAGCGGTGGAACTGAGCACCAACCGCTTA
AACACACGTCACACTTCC
CATGTCAGACACGAGAGAATTTAGACATTGTTTTGGAGAATCTACATTATAGAAATCAATGCTGAGACTC
ACATCTACATCATGTGTTATGATCAGCAGTATATGATATAATATTGATATTCATCAGTGTATTTACCCGC
AGGTGAGTGTTATGGGCAGTAATATATGATATAATATTGATATTCATCAGTGTATTTACCCGCAGGTGTG
TGTTATGATCAGCAGTATATGATATAATATTGATATTCATCAGTGTATTTACCCGCAGGTGAGTGTTATG
GGCAGTAATATATGATATAATATTGATATTCATCAGTGTATTTACCAGCAGGTGAGTGTTATGATCAGCA
GTATATGATATAATATTGATATTCATCAGTGTATTTACCCGCAGGTGAGTGTTATGGGCAGTAATATATG
ATATAATATTGATATTCATCAGTGTATTTACCCGCAGGTGAGTGTTATGGGCAGTAATATATGATATAAT
ATTGATATTCATCAGTGTATTTACCAGCAGGTGAGTGTTATGATCAGCAGTATATGATATAATATTGATA
TTCATCAGTGTATTTACCAGCAGGTGAGTGTTATGGGCAGTAATATATGATATAATATTGATATTCATCA
GTGTATTTACCAGCAGGTGAGTGTTATGATCAGCAGTATATGATATAATATTGATATTCATCAGTGTATT
TACCCGCAGGTGTGTGTTATGATCAGCAGTATATGATATAATATTGATATTCATCAGTGTATTTACCCGC
AGGTGAGTGTTATGGGCAGCAGGAGCGCGCAGGTCAGCAGTACGGCACCGCAGCACAGTATCTTTACGGG
TCTTCTCGCCATCTGATTCACCGCCTGCCGGAGCTCAGCCCGAACCCGCCGCAGCATCACCGACCGCTAC
AGACAGCAGCTCATCCCGGTGAACCGAACACAGCTGAACAGGAGCGGTGGAACTGAGCACCAACCGCTTA
AACACACGTCACACTTCC
