ZFLNCG00725
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1648856 | brain | normal | 63.49 |
| SRR519752 | 7 dpf | VD3 treatment | 36.63 |
| SRR891504 | liver | normal | 30.42 |
| SRR1299125 | caudal fin | Half day time after treatment | 30.39 |
| SRR1299126 | caudal fin | One day time after treatment | 27.72 |
| SRR519732 | 7 dpf | FETOH treatment | 27.29 |
| SRR1005530 | embryo | RPL11 morpholino | 26.33 |
| SRR658539 | bud | Gata5/6 morphant | 25.81 |
| SRR801556 | embryo | RPS19 and p53 morpholino | 25.61 |
| SRR658537 | bud | Gata6 morphant | 25.08 |
Express in tissues
Correlated coding gene
Download
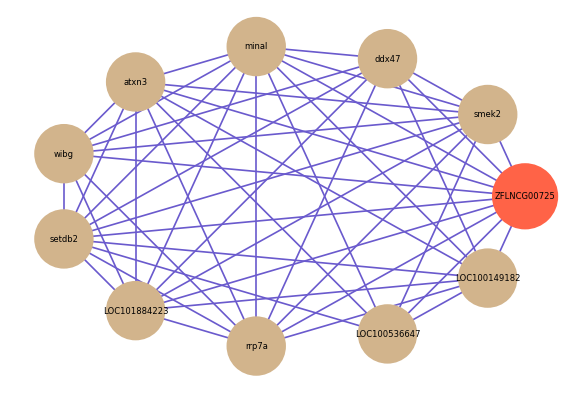
| gene | correlation coefficent |
|---|---|
| smek2 | 0.72 |
| ddx47 | 0.71 |
| minal | 0.70 |
| LOC100149182 | 0.70 |
| atxn3 | 0.68 |
| wibg | 0.66 |
| setdb2 | 0.65 |
| LOC101884223 | 0.64 |
| rrp7a | 0.64 |
| LOC100536647 | 0.64 |
Gene Ontology
Download
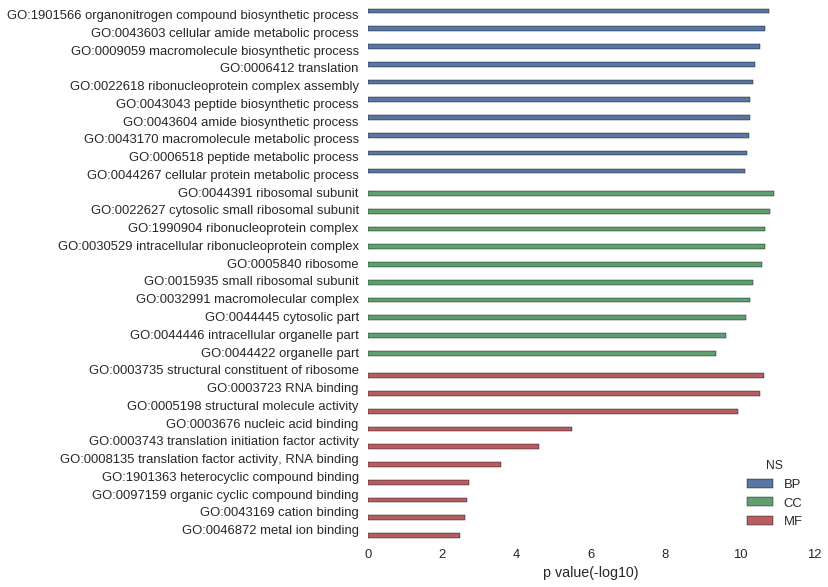
| GO | P value |
|---|---|
| GO:1901566 | 1.68e-11 |
| GO:0043603 | 2.13e-11 |
| GO:0009059 | 2.87e-11 |
| GO:0006412 | 3.79e-11 |
| GO:0022618 | 4.51e-11 |
| GO:0043043 | 5.31e-11 |
| GO:0043604 | 5.40e-11 |
| GO:0043170 | 5.51e-11 |
| GO:0006518 | 6.27e-11 |
| GO:0044267 | 7.35e-11 |
| GO:0044391 | 1.17e-11 |
| GO:0022627 | 1.57e-11 |
| GO:1990904 | 2.13e-11 |
| GO:0030529 | 2.13e-11 |
| GO:0005840 | 2.59e-11 |
| GO:0015935 | 4.33e-11 |
| GO:0032991 | 5.45e-11 |
| GO:0044445 | 6.61e-11 |
| GO:0044446 | 2.42e-10 |
| GO:0044422 | 4.29e-10 |
| GO:0003735 | 2.24e-11 |
| GO:0003723 | 2.91e-11 |
| GO:0005198 | 1.12e-10 |
| GO:0003676 | 3.25e-06 |
| GO:0003743 | 2.44e-05 |
| GO:0008135 | 2.64e-04 |
| GO:1901363 | 1.94e-03 |
| GO:0097159 | 2.11e-03 |
| GO:0043169 | 2.39e-03 |
| GO:0046872 | 3.36e-03 |
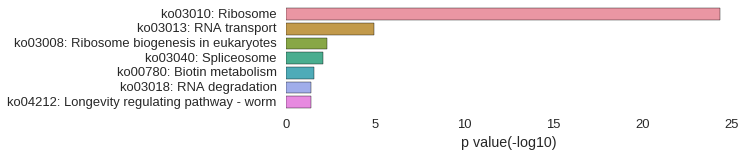
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00725
CTCTCTTAGTGTTTGGGTTAATTATTAATGCCCATTCAGAACTCTGCAGCTCTGGGTATTCTGAGCTGGA
CAAACACAGCACAGCAGCTCATTCAGCTTCATTTACAGCTTTATTCCACACACACACACACACACACACA
CACACACACACACACACACACCAGTGTGTAGACTATGAGTGTCTGACAAGAGCTGGAGTCACGTCTGCTT
GCACTGTATGAAGCAGAGGCCGAGGTGGTCCACGCCATCCCTATAACACACACACGCACACACACGCACA
CATGCACACACACACGCACACACACACACACGCACACACAAAGCATTACTTTGTAAGAAGAGTAGTGTGT
GTAAGTTTATAGTGTCTGTATGTGTAGTGTGTGTGTGTGTGTGTGTGTGTGTGTCATACACTCTGCTTCC
GGGCAGGATGACGATGCTCAGGCAGTTTGCCTCCAGCGCGCGATACACTTTCCCCACTCTTCTGTCCAGT
TTCCGGATCCTGCGCTCCAGCAGCCGCTCCTGCGCGTCACATGACACAAATCAACCAATCATATATCAAG
GACTGCGTTTCTGCATCTATTTGATTGACAGCTGTCACTCACATCACACTCCTCCCCCGGCTGTGCATTC
GAGATGGTTTCAGTAGTGGGCGGTTCAGGGGGCGTCCACGAGCTCATGTGGGGCGGGGTCAGAGCAGGAC
ATGCCCACAGCTTCCTGCCGCCAGTTAAGAGGCCGACGCCCACTGCAGCCTGAGCACTG
CTCTCTTAGTGTTTGGGTTAATTATTAATGCCCATTCAGAACTCTGCAGCTCTGGGTATTCTGAGCTGGA
CAAACACAGCACAGCAGCTCATTCAGCTTCATTTACAGCTTTATTCCACACACACACACACACACACACA
CACACACACACACACACACACCAGTGTGTAGACTATGAGTGTCTGACAAGAGCTGGAGTCACGTCTGCTT
GCACTGTATGAAGCAGAGGCCGAGGTGGTCCACGCCATCCCTATAACACACACACGCACACACACGCACA
CATGCACACACACACGCACACACACACACACGCACACACAAAGCATTACTTTGTAAGAAGAGTAGTGTGT
GTAAGTTTATAGTGTCTGTATGTGTAGTGTGTGTGTGTGTGTGTGTGTGTGTGTCATACACTCTGCTTCC
GGGCAGGATGACGATGCTCAGGCAGTTTGCCTCCAGCGCGCGATACACTTTCCCCACTCTTCTGTCCAGT
TTCCGGATCCTGCGCTCCAGCAGCCGCTCCTGCGCGTCACATGACACAAATCAACCAATCATATATCAAG
GACTGCGTTTCTGCATCTATTTGATTGACAGCTGTCACTCACATCACACTCCTCCCCCGGCTGTGCATTC
GAGATGGTTTCAGTAGTGGGCGGTTCAGGGGGCGTCCACGAGCTCATGTGGGGCGGGGTCAGAGCAGGAC
ATGCCCACAGCTTCCTGCCGCCAGTTAAGAGGCCGACGCCCACTGCAGCCTGAGCACTG