ZFLNCG00728
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR891511 | brain | normal | 26.96 |
| SRR1565817 | brain | normal | 15.54 |
| SRR1565809 | brain | normal | 14.54 |
| SRR1565811 | brain | normal | 14.27 |
| SRR1565819 | brain | normal | 11.09 |
| SRR1565805 | brain | normal | 10.32 |
| SRR1647683 | spleen | SVCV treatment | 9.02 |
| SRR1048061 | pineal gland | dark | 8.61 |
| SRR1565807 | brain | normal | 7.04 |
| SRR592703 | pineal gland | normal | 6.07 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
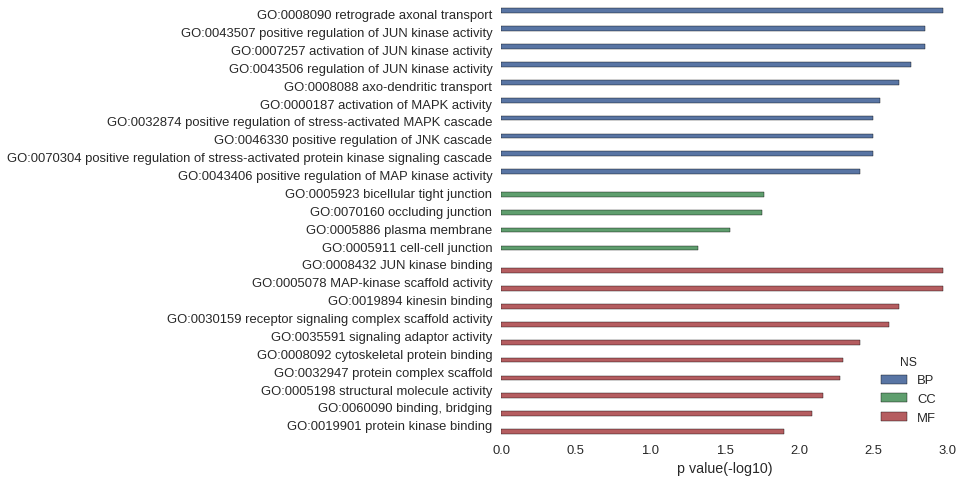
| GO | P value |
|---|---|
| GO:0008090 | 1.07e-03 |
| GO:0043507 | 1.42e-03 |
| GO:0007257 | 1.42e-03 |
| GO:0043506 | 1.78e-03 |
| GO:0008088 | 2.13e-03 |
| GO:0000187 | 2.84e-03 |
| GO:0032874 | 3.19e-03 |
| GO:0046330 | 3.19e-03 |
| GO:0070304 | 3.19e-03 |
| GO:0043406 | 3.90e-03 |
| GO:0005923 | 1.73e-02 |
| GO:0070160 | 1.76e-02 |
| GO:0005886 | 2.92e-02 |
| GO:0005911 | 4.77e-02 |
| GO:0008432 | 1.07e-03 |
| GO:0005078 | 1.07e-03 |
| GO:0019894 | 2.13e-03 |
| GO:0030159 | 2.49e-03 |
| GO:0035591 | 3.90e-03 |
| GO:0008092 | 5.05e-03 |
| GO:0032947 | 5.32e-03 |
| GO:0005198 | 6.89e-03 |
| GO:0060090 | 8.15e-03 |
| GO:0019901 | 1.27e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00728
GTTACCCTTAAGCCTAACTCTTGCCTCAGTGTCGCCCCCTGTGGCTGTGTCCGTTGACAGGAGCTGCGCT
GGAGTGTGTCTCAGTGTCGCCCCCTGCGGGTGCTGCTGACTCGAGTGCGTGCGGATGTGGCTCAGCTGGA
CTCTGTGCGTGACGAGCTGCAGGGGATCAGTGTGTCGCTGTCACTGCTGCAGGAACGCTTCACACTCCAG
CACTATCAGCAGCTGCAGCAGCGCATCTCCATCCTGCAGCAACACCTGCACAGCTGCTCCAGCCACCTGG
GTATTACACACACACACACATGCACTGACACCTGCCACACAAATGTATGCCCATTTTGGGAAATCACATT
AAAAACGACATGTGAATATTCATGTGTGTGTGTGTGTGTCCGCATGTGTGTGTTCAGGTTGTGGTCAGCT
GATCAGCATCAGTGCTCCGCTCACAGTCAGATCTTCAGGCTCTCGCTTCGGGTCGTGGATGATGGAGACT
TCCATTGAAAGTTCGGACAATCGCGTATGTCGAAGATCTGATTGTTCACTCTCACACACTCGACACCGAT
TGGACTCATTTTATTATATGAATCTTCACAACACCCTTAATGTGTGTGTGTGTGTGTGTGTGAGCAGGTG
TGGGTGATGGATGGATACTTAAAGGGCCGGCGTGTGTTGGAGT
GTTACCCTTAAGCCTAACTCTTGCCTCAGTGTCGCCCCCTGTGGCTGTGTCCGTTGACAGGAGCTGCGCT
GGAGTGTGTCTCAGTGTCGCCCCCTGCGGGTGCTGCTGACTCGAGTGCGTGCGGATGTGGCTCAGCTGGA
CTCTGTGCGTGACGAGCTGCAGGGGATCAGTGTGTCGCTGTCACTGCTGCAGGAACGCTTCACACTCCAG
CACTATCAGCAGCTGCAGCAGCGCATCTCCATCCTGCAGCAACACCTGCACAGCTGCTCCAGCCACCTGG
GTATTACACACACACACACATGCACTGACACCTGCCACACAAATGTATGCCCATTTTGGGAAATCACATT
AAAAACGACATGTGAATATTCATGTGTGTGTGTGTGTGTCCGCATGTGTGTGTTCAGGTTGTGGTCAGCT
GATCAGCATCAGTGCTCCGCTCACAGTCAGATCTTCAGGCTCTCGCTTCGGGTCGTGGATGATGGAGACT
TCCATTGAAAGTTCGGACAATCGCGTATGTCGAAGATCTGATTGTTCACTCTCACACACTCGACACCGAT
TGGACTCATTTTATTATATGAATCTTCACAACACCCTTAATGTGTGTGTGTGTGTGTGTGTGAGCAGGTG
TGGGTGATGGATGGATACTTAAAGGGCCGGCGTGTGTTGGAGT