ZFLNCG00737
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 362.56 |
| SRR891504 | liver | normal | 333.30 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 228.02 |
| SRR519732 | 7 dpf | FETOH treatment | 144.62 |
| SRR519727 | 6 dpf | FETOH treatment | 115.35 |
| SRR519752 | 7 dpf | VD3 treatment | 114.82 |
| SRR1035239 | liver | transgenic mCherry control | 99.89 |
| SRR519747 | 6 dpf | VD3 treatment | 99.35 |
| SRR891510 | muscle | normal | 98.24 |
| SRR891495 | heart | normal | 87.34 |
Express in tissues
Correlated coding gene
Download
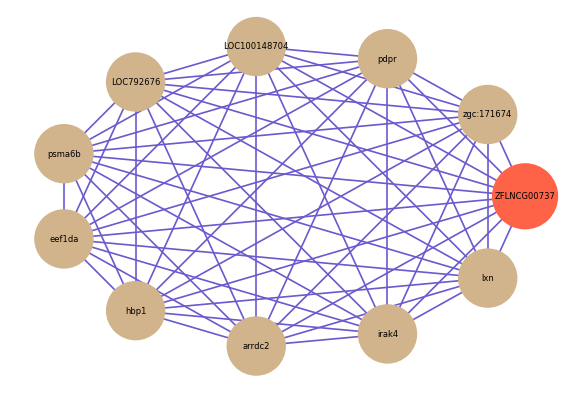
| gene | correlation coefficent |
|---|---|
| zgc:171674 | 0.74 |
| pdpr | 0.71 |
| LOC100148704 | 0.70 |
| LOC792676 | 0.69 |
| psma6b | 0.68 |
| eef1da | 0.68 |
| hbp1 | 0.68 |
| arrdc2 | 0.68 |
| irak4 | 0.67 |
| lxn | 0.67 |
Gene Ontology
Download
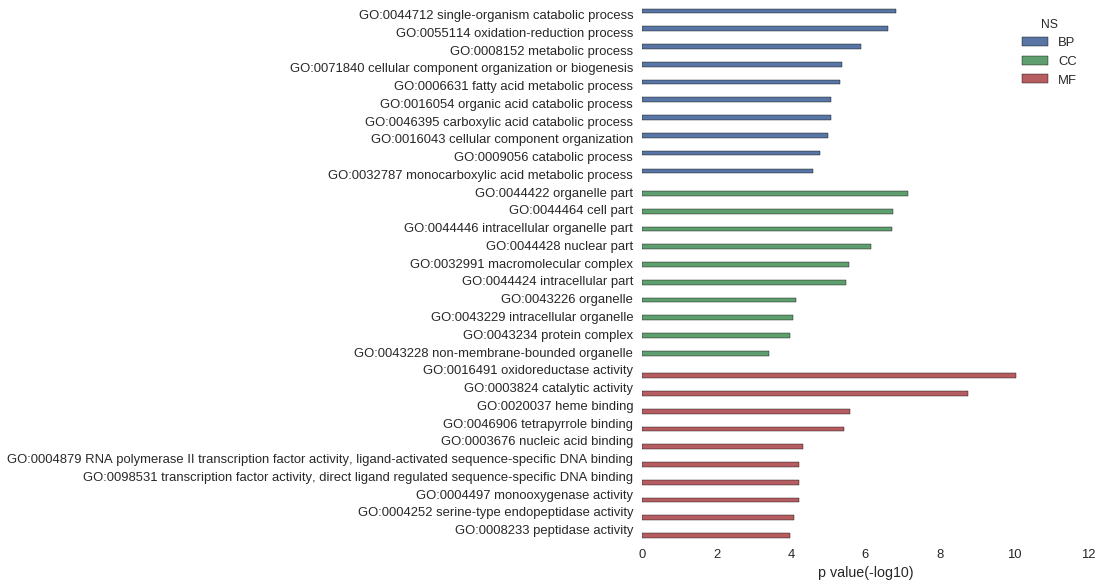
| GO | P value |
|---|---|
| GO:0044712 | 1.48e-07 |
| GO:0055114 | 2.54e-07 |
| GO:0008152 | 1.34e-06 |
| GO:0071840 | 4.18e-06 |
| GO:0006631 | 4.72e-06 |
| GO:0016054 | 8.21e-06 |
| GO:0046395 | 8.21e-06 |
| GO:0016043 | 1.02e-05 |
| GO:0009056 | 1.64e-05 |
| GO:0032787 | 2.54e-05 |
| GO:0044422 | 7.27e-08 |
| GO:0044464 | 1.83e-07 |
| GO:0044446 | 1.90e-07 |
| GO:0044428 | 7.30e-07 |
| GO:0032991 | 2.79e-06 |
| GO:0044424 | 3.24e-06 |
| GO:0043226 | 7.59e-05 |
| GO:0043229 | 8.99e-05 |
| GO:0043234 | 1.05e-04 |
| GO:0043228 | 3.98e-04 |
| GO:0016491 | 8.88e-11 |
| GO:0003824 | 1.72e-09 |
| GO:0020037 | 2.55e-06 |
| GO:0046906 | 3.88e-06 |
| GO:0003676 | 4.66e-05 |
| GO:0004879 | 6.28e-05 |
| GO:0098531 | 6.28e-05 |
| GO:0004497 | 6.31e-05 |
| GO:0004252 | 8.17e-05 |
| GO:0008233 | 1.07e-04 |
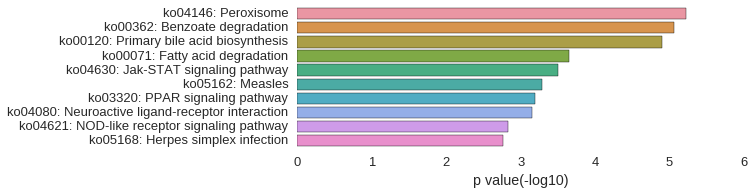
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00737
GCTGGCTCAGCTGCTCCAGGTAGCGTCCGCTGAGGGACATGTTCATCTCCAGCACTCGAATGCGGTTGCT
GAGCCTCATGAAGACCGACTCCTTCTGGTTGCCACTGTGGGCTGCTGTGCTGGCGTTCTGGGGGTCGGCG
AGGTCCAGCGTAGCGTCCGGTGGCTCCACATCACTGCTGTCCTCCGGTGGAGTGTCCAGTGAAGCGTCCG
CAGGCTCGGGGCTGTCCAGTGGAGCATCTGCTGGGGCGGCGGGGCCAGAAGGGGCCCCCTGAGAGCTGCT
GGAGCTTGGCTCAGTGTCCTCCTCAGAGGTGTGTGTGTCTGCAGTGCACTCGGAAACACACCCCGGCTCC
AGAGGCTCCGTCACACTGAAGGACGCACCGGACTCCAGCTGTGCGTCAGATGAGGGGTGTGTGTGTATGT
GTGTGAGCGGTGTCGGAGTGGGGCTGATGACCTCTGGAGCGCTGCACCGTTGCAGCAGATACTCCTGTAG
AGTGTGTGTATGCCAGTACTTGTCGTGCAGAGTGTGTGTGCGCCCGTCCTGCAGAGTGTGTGTGTGCTCA
TGCACATCCGCATCTGGCTCATCCTCAGCTTCGGGCAGCAGTGTGACGATCTGCTGTTCTTCAGGAGCTA
CCGACAGCGACTCTGGAGAAGCACTGACCGCTGCGGAGACACCCGACTGACCCACTGACCT
GCTGGCTCAGCTGCTCCAGGTAGCGTCCGCTGAGGGACATGTTCATCTCCAGCACTCGAATGCGGTTGCT
GAGCCTCATGAAGACCGACTCCTTCTGGTTGCCACTGTGGGCTGCTGTGCTGGCGTTCTGGGGGTCGGCG
AGGTCCAGCGTAGCGTCCGGTGGCTCCACATCACTGCTGTCCTCCGGTGGAGTGTCCAGTGAAGCGTCCG
CAGGCTCGGGGCTGTCCAGTGGAGCATCTGCTGGGGCGGCGGGGCCAGAAGGGGCCCCCTGAGAGCTGCT
GGAGCTTGGCTCAGTGTCCTCCTCAGAGGTGTGTGTGTCTGCAGTGCACTCGGAAACACACCCCGGCTCC
AGAGGCTCCGTCACACTGAAGGACGCACCGGACTCCAGCTGTGCGTCAGATGAGGGGTGTGTGTGTATGT
GTGTGAGCGGTGTCGGAGTGGGGCTGATGACCTCTGGAGCGCTGCACCGTTGCAGCAGATACTCCTGTAG
AGTGTGTGTATGCCAGTACTTGTCGTGCAGAGTGTGTGTGCGCCCGTCCTGCAGAGTGTGTGTGTGCTCA
TGCACATCCGCATCTGGCTCATCCTCAGCTTCGGGCAGCAGTGTGACGATCTGCTGTTCTTCAGGAGCTA
CCGACAGCGACTCTGGAGAAGCACTGACCGCTGCGGAGACACCCGACTGACCCACTGACCT