ZFLNCG00746
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647679 | head kidney | normal | 139.09 |
| SRR1647680 | head kidney | SVCV treatment | 127.98 |
| SRR1647681 | head kidney | SVCV treatment | 118.26 |
| ERR023146 | head kidney | normal | 37.57 |
| SRR1534349 | larvae | DMSO vehicle | 11.62 |
| SRR592702 | pineal gland | normal | 10.60 |
| SRR535847 | larvae | normal | 6.15 |
| SRR1167756 | embryo | mpeg1 morpholino | 5.67 |
| SRR1291417 | 5 dpi | injected marinum | 5.25 |
| SRR1534350 | larvae | 5 ug/l BDE47 treatment | 5.10 |
Express in tissues
Correlated coding gene
Download
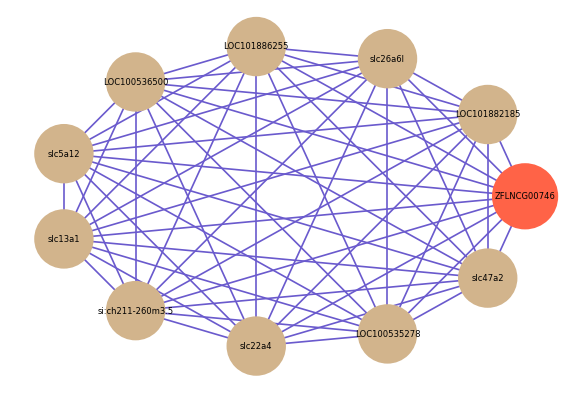
| gene | correlation coefficent |
|---|---|
| LOC101882185 | 0.76 |
| slc26a6l | 0.69 |
| LOC101886255 | 0.68 |
| LOC100536500 | 0.66 |
| slc5a12 | 0.65 |
| slc13a1 | 0.65 |
| si:ch211-260m3.5 | 0.65 |
| slc22a4 | 0.65 |
| LOC100535278 | 0.63 |
| slc47a2 | 0.63 |
Gene Ontology
Download
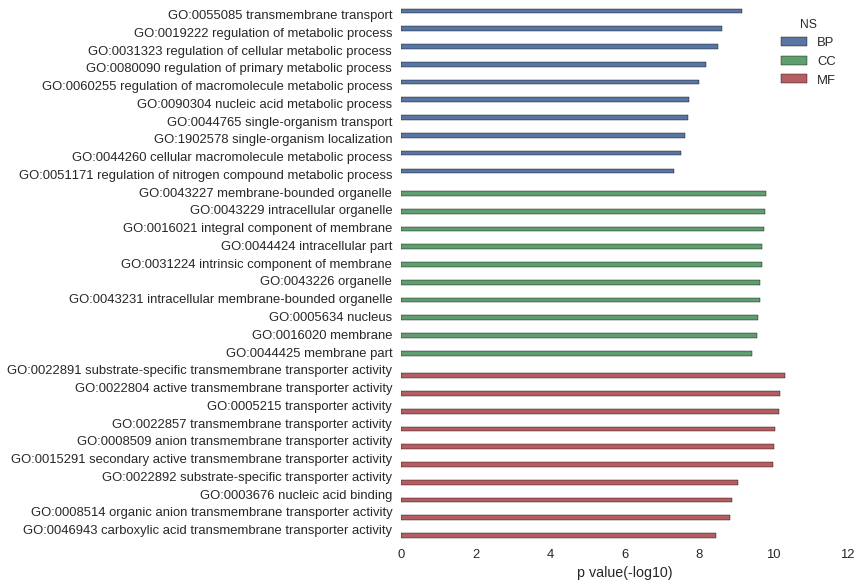
| GO | P value |
|---|---|
| GO:0055085 | 6.90e-10 |
| GO:0019222 | 2.33e-09 |
| GO:0031323 | 3.02e-09 |
| GO:0080090 | 6.62e-09 |
| GO:0060255 | 9.99e-09 |
| GO:0090304 | 1.87e-08 |
| GO:0044765 | 1.93e-08 |
| GO:1902578 | 2.35e-08 |
| GO:0044260 | 2.95e-08 |
| GO:0051171 | 4.72e-08 |
| GO:0043227 | 1.53e-10 |
| GO:0043229 | 1.70e-10 |
| GO:0016021 | 1.81e-10 |
| GO:0044424 | 1.99e-10 |
| GO:0031224 | 2.07e-10 |
| GO:0043226 | 2.24e-10 |
| GO:0043231 | 2.28e-10 |
| GO:0005634 | 2.60e-10 |
| GO:0016020 | 2.71e-10 |
| GO:0044425 | 3.77e-10 |
| GO:0022891 | 4.84e-11 |
| GO:0022804 | 6.81e-11 |
| GO:0005215 | 6.83e-11 |
| GO:0022857 | 8.93e-11 |
| GO:0008509 | 9.61e-11 |
| GO:0015291 | 1.02e-10 |
| GO:0022892 | 9.14e-10 |
| GO:0003676 | 1.28e-09 |
| GO:0008514 | 1.44e-09 |
| GO:0046943 | 3.43e-09 |
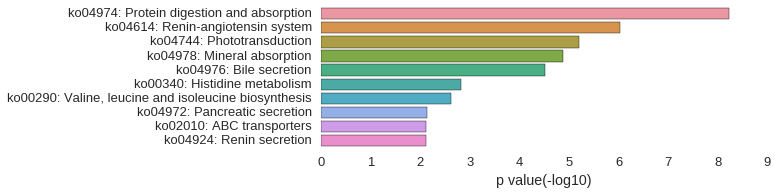
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00746
CCACACTTCCACCTGCACACTCTCTGAGCAGGTCTTCAGCTCTCTGCCGGACATCTGAAGGCAGACTTTG
GTCCTCCAGAAGAGCTGGATATGAGCAAGCAGCCAGAACCTAGGAGAGAAACACGGAGCGTTACAGACCA
CGCTTTCCTGGAGGATTCCTTCAAAGACATCTTTAAGGAAATCATTTACCTGCCGAACAAAGGTGATGGG
CTTTATTCCACCGCCGTGGAGATCTCCTGAAGACAGATCCAGCACCTCACTGAAGATCTTTCTCTCACCC
TGAAAACAACATGACACCTGTTCTTCAGTCACACACACTCCTGAGAGGAAAGTGTGTGTGCTGGCTTTAC
TGGTGCATGAAGAAACTTTACATTGTGCTAAAGTACAGACAGATAGATAGATAGATAGATAGATAGATAG
ATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGAT
AGATAGATAGATAGATAGACAGACATAAGCAAACACACACACAAACACAAACACACACACCTGCTGGATC
TCCCTCTGTATCTCCGCTGCTCTGCTCTGCAAGATGTCCTTCTCGGAGCGGTGGATCTGCAGGACAGACA
TCATTGTTCCTCTGGAGACTCGAACACCCATAATCAGCTCAGCGAGGCTGTA
CCACACTTCCACCTGCACACTCTCTGAGCAGGTCTTCAGCTCTCTGCCGGACATCTGAAGGCAGACTTTG
GTCCTCCAGAAGAGCTGGATATGAGCAAGCAGCCAGAACCTAGGAGAGAAACACGGAGCGTTACAGACCA
CGCTTTCCTGGAGGATTCCTTCAAAGACATCTTTAAGGAAATCATTTACCTGCCGAACAAAGGTGATGGG
CTTTATTCCACCGCCGTGGAGATCTCCTGAAGACAGATCCAGCACCTCACTGAAGATCTTTCTCTCACCC
TGAAAACAACATGACACCTGTTCTTCAGTCACACACACTCCTGAGAGGAAAGTGTGTGTGCTGGCTTTAC
TGGTGCATGAAGAAACTTTACATTGTGCTAAAGTACAGACAGATAGATAGATAGATAGATAGATAGATAG
ATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGAT
AGATAGATAGATAGATAGACAGACATAAGCAAACACACACACAAACACAAACACACACACCTGCTGGATC
TCCCTCTGTATCTCCGCTGCTCTGCTCTGCAAGATGTCCTTCTCGGAGCGGTGGATCTGCAGGACAGACA
TCATTGTTCCTCTGGAGACTCGAACACCCATAATCAGCTCAGCGAGGCTGTA