ZFLNCG00753
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR748488 | sperm | normal | 1.14 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 0.86 |
| SRR800049 | sphere stage | control treatment | 0.69 |
| SRR748490 | 1 kcell | normal | 0.69 |
| SRR800043 | sphere stage | normal | 0.63 |
| SRR372793 | shield | normal | 0.43 |
| SRR372791 | dome | normal | 0.43 |
| SRR1021215 | sphere stage | Mzeomesa | 0.42 |
| SRR748491 | germ ring | normal | 0.41 |
| SRR1562533 | testis | normal | 0.40 |
Express in tissues
Correlated coding gene
Download
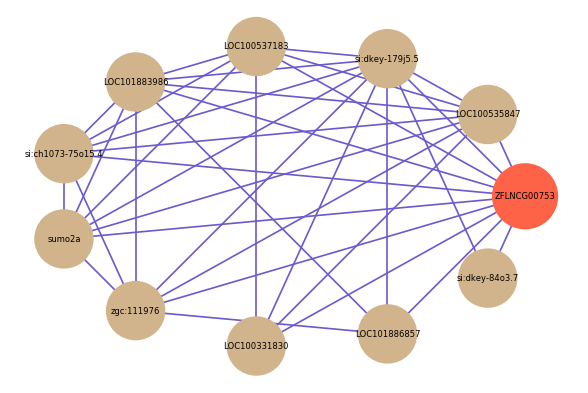
| gene | correlation coefficent |
|---|---|
| LOC100535847 | 0.60 |
| si:dkey-179j5.5 | 0.57 |
| LOC100537183 | 0.56 |
| LOC101883986 | 0.56 |
| si:ch1073-75o15.4 | 0.55 |
| sumo2a | 0.55 |
| zgc:111976 | 0.54 |
| LOC100331830 | 0.54 |
| LOC101886857 | 0.54 |
| si:dkey-84o3.7 | 0.54 |
Gene Ontology
Download
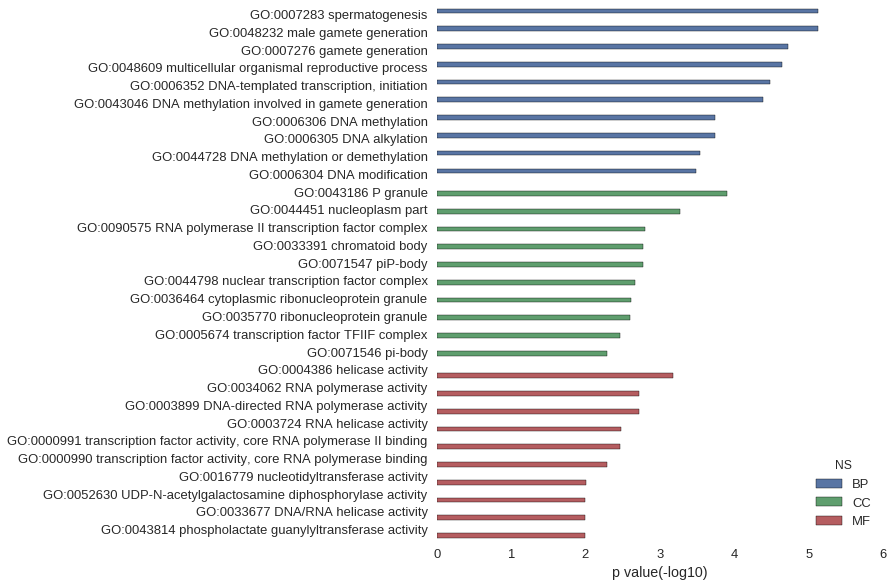
| GO | P value |
|---|---|
| GO:0007283 | 7.55e-06 |
| GO:0048232 | 7.55e-06 |
| GO:0007276 | 1.90e-05 |
| GO:0048609 | 2.30e-05 |
| GO:0006352 | 3.26e-05 |
| GO:0043046 | 4.16e-05 |
| GO:0006306 | 1.82e-04 |
| GO:0006305 | 1.82e-04 |
| GO:0044728 | 2.89e-04 |
| GO:0006304 | 3.30e-04 |
| GO:0043186 | 1.24e-04 |
| GO:0044451 | 5.35e-04 |
| GO:0090575 | 1.60e-03 |
| GO:0033391 | 1.71e-03 |
| GO:0071547 | 1.71e-03 |
| GO:0044798 | 2.19e-03 |
| GO:0036464 | 2.41e-03 |
| GO:0035770 | 2.52e-03 |
| GO:0005674 | 3.41e-03 |
| GO:0071546 | 5.11e-03 |
| GO:0004386 | 6.70e-04 |
| GO:0034062 | 1.89e-03 |
| GO:0003899 | 1.89e-03 |
| GO:0003724 | 3.38e-03 |
| GO:0000991 | 3.41e-03 |
| GO:0000990 | 5.11e-03 |
| GO:0016779 | 9.97e-03 |
| GO:0052630 | 1.02e-02 |
| GO:0033677 | 1.02e-02 |
| GO:0043814 | 1.02e-02 |
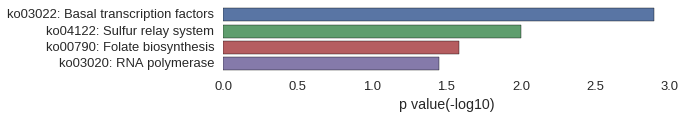
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00753
GTGATTGTGATAGTGTTTTATTGTCGTCTCTCAGGTTTTATCTCTGCTTTAATGCCCTTTAAAAATAAAT
AAATGAAAAACAAAGCAGCTGCTTGCCATCACGACAGCAAAAAGATCCAACGGACAGCCGATCGCTTTCA
CTTCAAAATGGCGGAATCCGGGGCTGTTGCGGGGCGCTGAGGTTGCAATGGAAAGCTCTATTGAGTGTCG
CCTCTTGGTCATCCTCATAGAAATATACATATATCTGTATATATCTATGGGTCATTCTAAGCTCTTTGAT
AGAGCCCATTAACGGTGAGAGGGTATTTGCTATGGAAACACTCAGGTTGACATCAATGCGGACTCCAAAC
GCGGAAGCAAAAGGTGAGATTTTCACCGATGATTATCAAAACAAACACATTTGTTTCCATATACTTCTAG
AATAACTTTGTGAGCTGGCAAAGTAAACGATGTACATTTTGATATCTAGCAAACGTTTAAAGTGACATAA
ATAGGCTGAACAGAACTTAAGTCCGTTCATTTAACGTTTTCCGTCTTTAATACAAGCGAAAAACATCCCG
TCCAACAATATTTTGACATGTTGTCTTCCACACTCGGTCACGAAAAGGAAACCGAACTTAGTGGTAGCAT
GTAAACAAGAACTCGAACTATCCAGGTCAAAATAAAGGAGCTGCTGAGAGATGTGTGTGTGTGTTTGTGT
GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTAGCTGTGTGTCTTGATTTTGGAGGGCTCGAGAGAGC
GAGAAAAGGAGGACAAAGGGAAACATGGACGCTTGCCATACCTCGCTAGCATCATTATGGGACTATCAAG
TGTCCGTTTCAGCGTTTGACCATCGCCACTGGCAGGTTACTGCATTTCTAGCAGATTTTACCTTCCGTCT
TACCTGTAACCCGAGCCAATATTTTGTGCAGTCTGCATTCCTGGAGACACTGTAAAAGAGCACGATATTA
GAGCTCTCTGCCAAACCTCTCAATTGAGCAGATTTAATTTTAACATTTCTGCAGGACCCCCAGCCGCTTT
CTGCTGAGGTCGCCTGAGTTTGATATAACAGGACGTATTTTGGTTTCTGTAATTGGCTCCATATGATCGC
GATTGTCTGTGGTCGTAGTTCCTAATACATGATTTGAAATGCGCATCTGTTCGCGTTACTTTTTTTTTGG
GCGGGGGTTCCAAC
GTGATTGTGATAGTGTTTTATTGTCGTCTCTCAGGTTTTATCTCTGCTTTAATGCCCTTTAAAAATAAAT
AAATGAAAAACAAAGCAGCTGCTTGCCATCACGACAGCAAAAAGATCCAACGGACAGCCGATCGCTTTCA
CTTCAAAATGGCGGAATCCGGGGCTGTTGCGGGGCGCTGAGGTTGCAATGGAAAGCTCTATTGAGTGTCG
CCTCTTGGTCATCCTCATAGAAATATACATATATCTGTATATATCTATGGGTCATTCTAAGCTCTTTGAT
AGAGCCCATTAACGGTGAGAGGGTATTTGCTATGGAAACACTCAGGTTGACATCAATGCGGACTCCAAAC
GCGGAAGCAAAAGGTGAGATTTTCACCGATGATTATCAAAACAAACACATTTGTTTCCATATACTTCTAG
AATAACTTTGTGAGCTGGCAAAGTAAACGATGTACATTTTGATATCTAGCAAACGTTTAAAGTGACATAA
ATAGGCTGAACAGAACTTAAGTCCGTTCATTTAACGTTTTCCGTCTTTAATACAAGCGAAAAACATCCCG
TCCAACAATATTTTGACATGTTGTCTTCCACACTCGGTCACGAAAAGGAAACCGAACTTAGTGGTAGCAT
GTAAACAAGAACTCGAACTATCCAGGTCAAAATAAAGGAGCTGCTGAGAGATGTGTGTGTGTGTTTGTGT
GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTAGCTGTGTGTCTTGATTTTGGAGGGCTCGAGAGAGC
GAGAAAAGGAGGACAAAGGGAAACATGGACGCTTGCCATACCTCGCTAGCATCATTATGGGACTATCAAG
TGTCCGTTTCAGCGTTTGACCATCGCCACTGGCAGGTTACTGCATTTCTAGCAGATTTTACCTTCCGTCT
TACCTGTAACCCGAGCCAATATTTTGTGCAGTCTGCATTCCTGGAGACACTGTAAAAGAGCACGATATTA
GAGCTCTCTGCCAAACCTCTCAATTGAGCAGATTTAATTTTAACATTTCTGCAGGACCCCCAGCCGCTTT
CTGCTGAGGTCGCCTGAGTTTGATATAACAGGACGTATTTTGGTTTCTGTAATTGGCTCCATATGATCGC
GATTGTCTGTGGTCGTAGTTCCTAATACATGATTTGAAATGCGCATCTGTTCGCGTTACTTTTTTTTTGG
GCGGGGGTTCCAAC