ZFLNCG00845
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1565819 | brain | normal | 15.99 |
| SRR1565817 | brain | normal | 15.66 |
| SRR1565815 | brain | normal | 14.45 |
| SRR1035978 | 13 hpf | rx3-/- | 13.90 |
| SRR516122 | skin | male and 3.5 year | 13.63 |
| SRR527833 | 5 dpf | normal | 12.79 |
| SRR658535 | bud | Gata5 morphant | 12.45 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 11.94 |
| SRR1565811 | brain | normal | 11.80 |
| ERR023144 | brain | normal | 11.48 |
Express in tissues
Correlated coding gene
Download
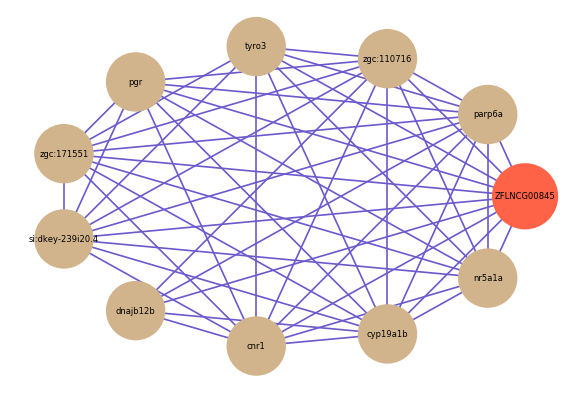
| gene | correlation coefficent |
|---|---|
| parp6a | 0.55 |
| zgc:110716 | 0.55 |
| tyro3 | 0.54 |
| pgr | 0.54 |
| zgc:171551 | 0.54 |
| si:dkey-239i20.4 | 0.54 |
| dnajb12b | 0.53 |
| cnr1 | 0.53 |
| cyp19a1b | 0.53 |
| nr5a1a | 0.53 |
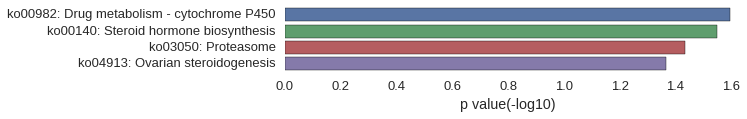
Gene Ontology
Download

| GO | P value |
|---|---|
| GO:0030728 | 1.70e-03 |
| GO:0043401 | 1.83e-03 |
| GO:0009755 | 2.18e-03 |
| GO:2000253 | 2.56e-03 |
| GO:0017144 | 2.56e-03 |
| GO:0060259 | 4.26e-03 |
| GO:0048520 | 6.80e-03 |
| GO:0032355 | 6.80e-03 |
| GO:0030325 | 7.65e-03 |
| GO:0021854 | 8.50e-03 |
| GO:0042734 | 9.34e-03 |
| GO:0090575 | 2.95e-02 |
| GO:0044798 | 3.44e-02 |
| GO:0004949 | 1.70e-03 |
| GO:0003707 | 1.93e-03 |
| GO:0038023 | 2.37e-03 |
| GO:0004499 | 3.41e-03 |
| GO:0004497 | 3.93e-03 |
| GO:0004872 | 4.24e-03 |
| GO:0060089 | 4.24e-03 |
| GO:0004871 | 4.73e-03 |
| GO:0016705 | 6.50e-03 |
| GO:0016709 | 1.36e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00845
AGCAGTAAAAGTCAAAGCCCCTACTCACCCAAAAATGAACAGGACAGGAGTAAAGGTAATTAAGAAATAA
CACTGAAACCCATCAATGTCAGTGAAATGTTTTTCTGATGTCAGCGTTGTGTTTCAGATGACTCTAGTCC
TCCGAGAGATAAAGCTGCGTGGAGAAGAGGTATTCCAGGTTTAAAGAAAAAGCCACAAGATAAGAGGCCT
GCTGCTGGCGTCACATTTGGAGTACGACTGGATAACTGCCCTCCTGCGCAAACCAATAAAGTAAGTGATT
TAAACATGTCATTAGTTATGTTTTCATTAAAAAATGTGAATGAACTTTGTGCAAAACTGAAATCTCGCAT
AAAAGATCTG
AGCAGTAAAAGTCAAAGCCCCTACTCACCCAAAAATGAACAGGACAGGAGTAAAGGTAATTAAGAAATAA
CACTGAAACCCATCAATGTCAGTGAAATGTTTTTCTGATGTCAGCGTTGTGTTTCAGATGACTCTAGTCC
TCCGAGAGATAAAGCTGCGTGGAGAAGAGGTATTCCAGGTTTAAAGAAAAAGCCACAAGATAAGAGGCCT
GCTGCTGGCGTCACATTTGGAGTACGACTGGATAACTGCCCTCCTGCGCAAACCAATAAAGTAAGTGATT
TAAACATGTCATTAGTTATGTTTTCATTAAAAAATGTGAATGAACTTTGTGCAAAACTGAAATCTCGCAT
AAAAGATCTG