ZFLNCG01027
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800045 | muscle | normal | 554.17 |
| SRR800037 | egg | normal | 221.15 |
| SRR891495 | heart | normal | 193.94 |
| SRR516122 | skin | male and 3.5 year | 114.45 |
| SRR527835 | tail | normal | 106.72 |
| SRR1647679 | head kidney | normal | 95.29 |
| SRR527833 | 5 dpf | normal | 82.05 |
| SRR1647680 | head kidney | SVCV treatment | 66.99 |
| SRR1647681 | head kidney | SVCV treatment | 65.19 |
| SRR957181 | heart | normal | 61.52 |
Express in tissues
Correlated coding gene
Download
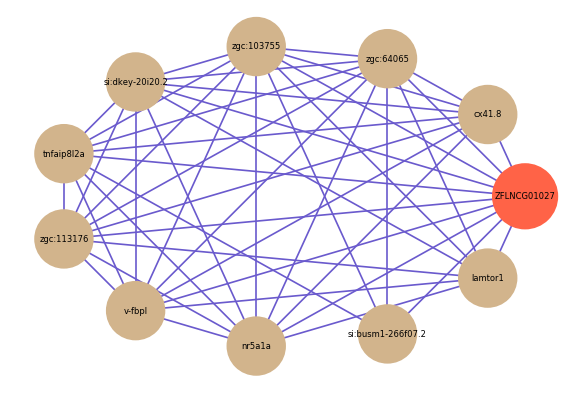
| gene | correlation coefficent |
|---|---|
| cx41.8 | 0.61 |
| zgc:64065 | 0.59 |
| zgc:103755 | 0.57 |
| si:dkey-20i20.2 | 0.57 |
| tnfaip8l2a | 0.57 |
| zgc:113176 | 0.56 |
| v-fbpl | 0.55 |
| si:busm1-266f07.2 | 0.53 |
| lamtor1 | 0.52 |
| nr5a1a | 0.52 |
Gene Ontology
Download
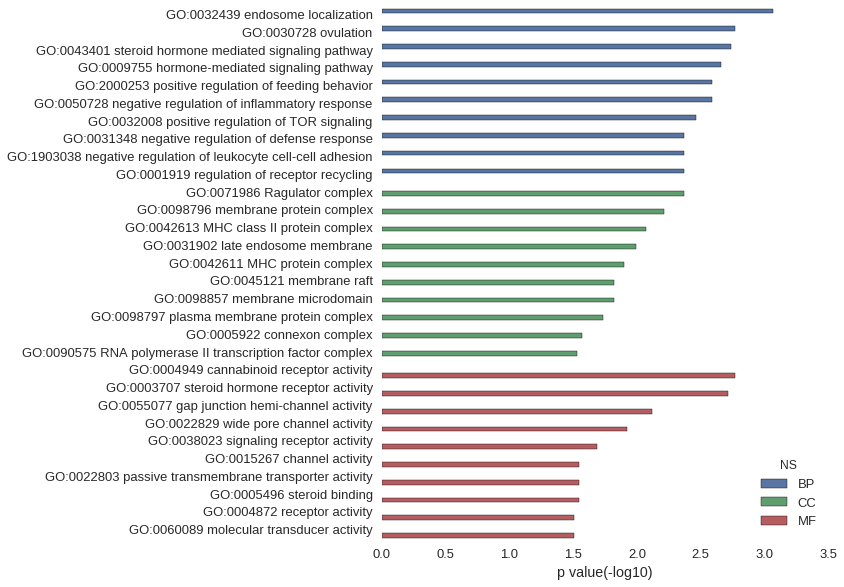
| GO | P value |
|---|---|
| GO:0032439 | 8.53e-04 |
| GO:0030728 | 1.70e-03 |
| GO:0043401 | 1.83e-03 |
| GO:0009755 | 2.18e-03 |
| GO:2000253 | 2.56e-03 |
| GO:0050728 | 2.56e-03 |
| GO:0032008 | 3.41e-03 |
| GO:0031348 | 4.26e-03 |
| GO:1903038 | 4.26e-03 |
| GO:0001919 | 4.26e-03 |
| GO:0071986 | 4.26e-03 |
| GO:0098796 | 6.13e-03 |
| GO:0042613 | 8.50e-03 |
| GO:0031902 | 1.02e-02 |
| GO:0042611 | 1.27e-02 |
| GO:0045121 | 1.52e-02 |
| GO:0098857 | 1.52e-02 |
| GO:0098797 | 1.83e-02 |
| GO:0005922 | 2.70e-02 |
| GO:0090575 | 2.95e-02 |
| GO:0004949 | 1.70e-03 |
| GO:0003707 | 1.93e-03 |
| GO:0055077 | 7.65e-03 |
| GO:0022829 | 1.19e-02 |
| GO:0038023 | 2.04e-02 |
| GO:0015267 | 2.83e-02 |
| GO:0022803 | 2.83e-02 |
| GO:0005496 | 2.86e-02 |
| GO:0004872 | 3.11e-02 |
| GO:0060089 | 3.11e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01027
TGGACTCCTGGGGTATCAGCATCTACAATGAAGCCTGTGAAAGCCTTGCTAGCAGGACATTTGGGATCGG
AATCTGTGCGGGCCAGCAGGAAGTACCTTTAAATGCAAAACAAAAGGGTTTATACTTGGCCTAAAACAAA
TTTATGAATTCATTGTAGGAATTTGAAATGCTTCTCCTACCAGTTTGCTTTTCCTCCATTAGTGATCCAC
ATTTTCTGACCATTGATCACGTAGTCATCTCCTTTCTTAACAGCCCGGGTCTTTATTCCAGCCACATCAG
AGCCTGCTCCTGGCTCAGTTACACAGTACGCCTGTCCACACACAGTACCATGTTAATGACAAAAAGTGTG
CAAGTACCATTATGAGAACACTTCAGAAATCATGAGATGGTTAGAACATTTGACACTTACACACATCAAT
GGTTCCTCTGTCATTCTGCCCAAATATTTCTTTCTCTGAGCATCATTACCAGCGATGATGACTGGCATTT
GCTGCATGGAGAAAATAAATAAACACCTCTAAAAATCACAGTAACTGTTTTAGACTAATCATAACTCCTC
CTCATGTTCACTACATAACAAAAATACAATGTGCTACGAGTCCTAACTACAAATTGCCATTGTTTTCAAA
TATTTATTACTTGTAAATACAATTTTTAAACTCCAGAGACTTACCCCCAGAGAGTTCGCCTCAATGGCTG
TCTGCACCCCAGTGCAACCATAGGCCAGCTCTTCTGTGATGAGACAGGCATCAAAAATGCCCAGACCCAT
TCCACCTAAAATATACCGCAACATCCAACATGTTCTAAAATTATTATCATAACTCTTACATCCATACTGA
AAAAAAAAAA
TGGACTCCTGGGGTATCAGCATCTACAATGAAGCCTGTGAAAGCCTTGCTAGCAGGACATTTGGGATCGG
AATCTGTGCGGGCCAGCAGGAAGTACCTTTAAATGCAAAACAAAAGGGTTTATACTTGGCCTAAAACAAA
TTTATGAATTCATTGTAGGAATTTGAAATGCTTCTCCTACCAGTTTGCTTTTCCTCCATTAGTGATCCAC
ATTTTCTGACCATTGATCACGTAGTCATCTCCTTTCTTAACAGCCCGGGTCTTTATTCCAGCCACATCAG
AGCCTGCTCCTGGCTCAGTTACACAGTACGCCTGTCCACACACAGTACCATGTTAATGACAAAAAGTGTG
CAAGTACCATTATGAGAACACTTCAGAAATCATGAGATGGTTAGAACATTTGACACTTACACACATCAAT
GGTTCCTCTGTCATTCTGCCCAAATATTTCTTTCTCTGAGCATCATTACCAGCGATGATGACTGGCATTT
GCTGCATGGAGAAAATAAATAAACACCTCTAAAAATCACAGTAACTGTTTTAGACTAATCATAACTCCTC
CTCATGTTCACTACATAACAAAAATACAATGTGCTACGAGTCCTAACTACAAATTGCCATTGTTTTCAAA
TATTTATTACTTGTAAATACAATTTTTAAACTCCAGAGACTTACCCCCAGAGAGTTCGCCTCAATGGCTG
TCTGCACCCCAGTGCAACCATAGGCCAGCTCTTCTGTGATGAGACAGGCATCAAAAATGCCCAGACCCAT
TCCACCTAAAATATACCGCAACATCCAACATGTTCTAAAATTATTATCATAACTCTTACATCCATACTGA
AAAAAAAAAA