ZFLNCG01029
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR527835 | tail | normal | 22.74 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 17.11 |
| SRR891504 | liver | normal | 15.13 |
| SRR527834 | head | normal | 14.48 |
| SRR527833 | 5 dpf | normal | 14.11 |
| SRR1035978 | 13 hpf | rx3-/- | 11.10 |
| SRR726542 | 5 dpf | infection with control | 10.88 |
| SRR658537 | bud | Gata6 morphant | 10.77 |
| SRR658539 | bud | Gata5/6 morphant | 8.52 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 8.39 |
Express in tissues
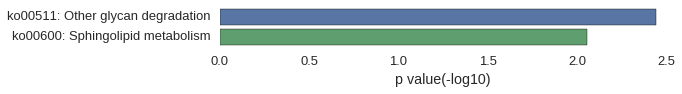
Gene Ontology
Download
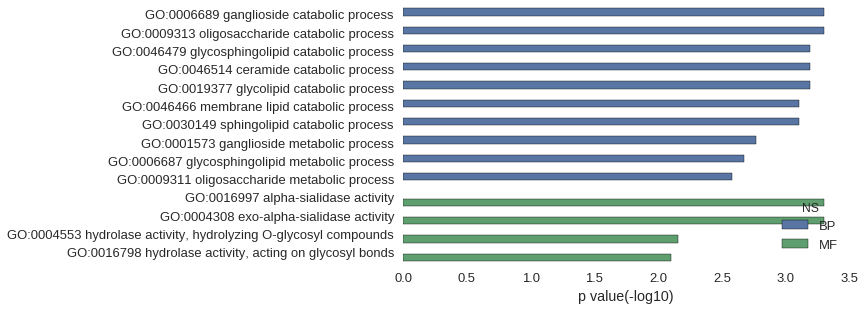
| GO | P value |
|---|---|
| GO:0006689 | 4.97e-04 |
| GO:0009313 | 4.97e-04 |
| GO:0046479 | 6.40e-04 |
| GO:0046514 | 6.40e-04 |
| GO:0019377 | 6.40e-04 |
| GO:0046466 | 7.82e-04 |
| GO:0030149 | 7.82e-04 |
| GO:0001573 | 1.71e-03 |
| GO:0006687 | 2.13e-03 |
| GO:0009311 | 2.63e-03 |
| GO:0016997 | 4.97e-04 |
| GO:0004308 | 4.97e-04 |
| GO:0004553 | 6.96e-03 |
| GO:0016798 | 7.96e-03 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01029
TATGTGCCTCCGACATGGACGTTTGAATGTGACGAGGACCTCGTGCACTACTTCTATGACCACATTGGAA
AGGAGGATGAAAATCTTGGCAGTGTGAAGCAATGTGTGACCAGTATTGACGTGTCTTCTAGTTCGGTATG
TGATGGCACAGTATCATTCTGCATTACACTGAATAGCAGAGAGCTGCAAACATATCAGATGTTTTCGACT
AGTAAGGGAGTTGCTCTGCTTGAAGGAGGACCCTAGTGGTGGAGCAAGCTGTCTGACAGATGGAGACACG
GAGACGTACTGGGAGAGCGATGGCATGCAGGGACAGCACTGGATCCGTCTGCACATGAGGAGAGGCACTG
TGGTCAAGTAAGGACGACTGGTCTGCTAGCGACTGTAATTAAAAAGGACTTTTTCCACACTGTACTTAAA
TCCAGTTGGTTCTAGATAATTTCACAAGTTATGAAATCAGGTCAGTCTGTTCAAACTTTTGTTTTAATGA
TTTA
TATGTGCCTCCGACATGGACGTTTGAATGTGACGAGGACCTCGTGCACTACTTCTATGACCACATTGGAA
AGGAGGATGAAAATCTTGGCAGTGTGAAGCAATGTGTGACCAGTATTGACGTGTCTTCTAGTTCGGTATG
TGATGGCACAGTATCATTCTGCATTACACTGAATAGCAGAGAGCTGCAAACATATCAGATGTTTTCGACT
AGTAAGGGAGTTGCTCTGCTTGAAGGAGGACCCTAGTGGTGGAGCAAGCTGTCTGACAGATGGAGACACG
GAGACGTACTGGGAGAGCGATGGCATGCAGGGACAGCACTGGATCCGTCTGCACATGAGGAGAGGCACTG
TGGTCAAGTAAGGACGACTGGTCTGCTAGCGACTGTAATTAAAAAGGACTTTTTCCACACTGTACTTAAA
TCCAGTTGGTTCTAGATAATTTCACAAGTTATGAAATCAGGTCAGTCTGTTCAAACTTTTGTTTTAATGA
TTTA