ZFLNCG01229
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 55.21 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 41.00 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 34.07 |
| SRR594769 | blood | normal | 28.57 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 20.49 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 17.94 |
| SRR594771 | endothelium | normal | 15.71 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 14.63 |
| SRR592698 | pineal gland | normal | 14.55 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 12.69 |
Express in tissues
Correlated coding gene
Download
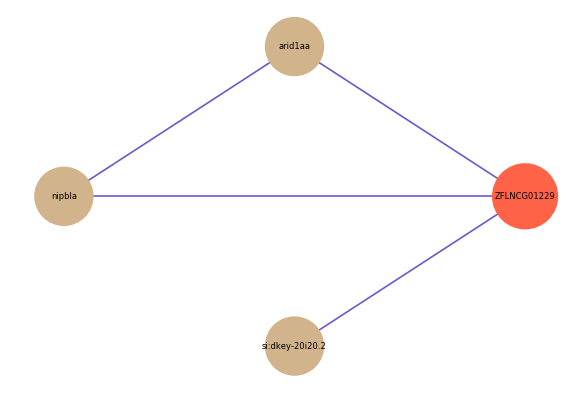
| gene | correlation coefficent |
|---|---|
| arid1aa | 0.54 |
| nipbla | 0.52 |
| si:dkey-20i20.2 | 0.50 |
Gene Ontology
Download
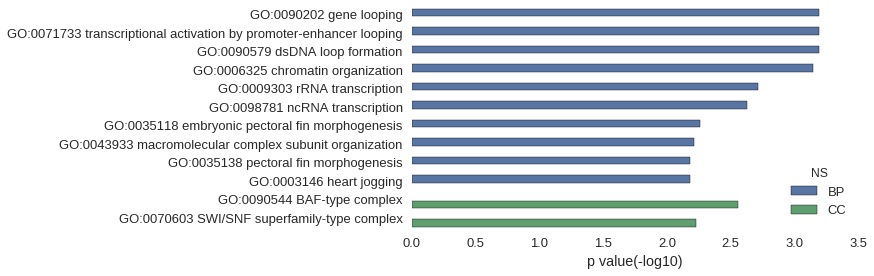
| GO | P value |
|---|---|
| GO:0090202 | 6.39e-04 |
| GO:0071733 | 6.39e-04 |
| GO:0090579 | 6.39e-04 |
| GO:0006325 | 7.16e-04 |
| GO:0009303 | 1.92e-03 |
| GO:0098781 | 2.34e-03 |
| GO:0035118 | 5.53e-03 |
| GO:0043933 | 6.16e-03 |
| GO:0035138 | 6.59e-03 |
| GO:0003146 | 6.59e-03 |
| GO:0090544 | 2.77e-03 |
| GO:0070603 | 5.96e-03 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01229
TTCTCCTCTAAAAGGTGAACATAGAGCCTTCGTGCCCTTTGACCTCTGAGCCTCTTCTGTAAGGTGACGG
TGGCCCATCGCATTGTCTGAAAGCGTCGGCGCCAATATAGAGCACGGTAATTTTTCTGAATGACAACAAT
GCACAGCAACAGGCGTCTGTACTGCCTCCTGTGACCAGCAGAGAGCAAAAACACAGTGACTACATTAGAG
TTTAGTACTTTAGTGTTTGTTTGTGTATATGCACGTGCCAGTGTGTTTACCGTGCTCTGTAGCCCAGTAT
ATGAGCTTGGATAATTGATGCTGCTTTGAGAACTTCTGTTTCTCTTTGTTTTTCTAACCGCAGCTCCAAA
CTCTCTCTC
TTCTCCTCTAAAAGGTGAACATAGAGCCTTCGTGCCCTTTGACCTCTGAGCCTCTTCTGTAAGGTGACGG
TGGCCCATCGCATTGTCTGAAAGCGTCGGCGCCAATATAGAGCACGGTAATTTTTCTGAATGACAACAAT
GCACAGCAACAGGCGTCTGTACTGCCTCCTGTGACCAGCAGAGAGCAAAAACACAGTGACTACATTAGAG
TTTAGTACTTTAGTGTTTGTTTGTGTATATGCACGTGCCAGTGTGTTTACCGTGCTCTGTAGCCCAGTAT
ATGAGCTTGGATAATTGATGCTGCTTTGAGAACTTCTGTTTCTCTTTGTTTTTCTAACCGCAGCTCCAAA
CTCTCTCTC

