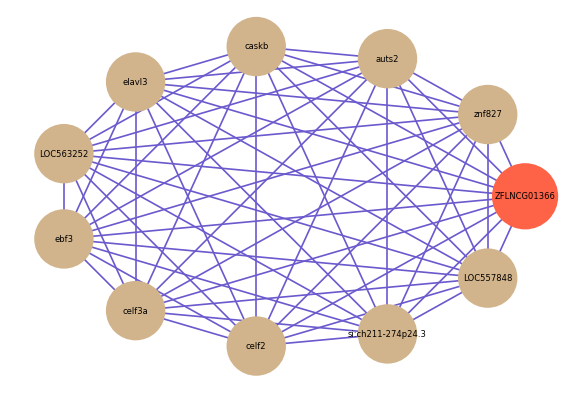
ZFLNCG01366
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR514028 | retina | control morpholino | 49.97 |
| SRR514030 | retina | id2a morpholino | 45.56 |
| ERR023144 | brain | normal | 37.57 |
| SRR1648855 | brain | normal | 9.90 |
| SRR891511 | brain | normal | 6.28 |
| ERR023145 | heart | normal | 6.16 |
| SRR1648854 | brain | normal | 5.74 |
| SRR726540 | 5 dpf | normal | 3.90 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 3.51 |
| SRR519717 | 2 dpf | FETOH treatment | 3.14 |
Express in tissues
Correlated coding gene
Download
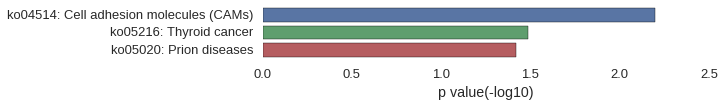
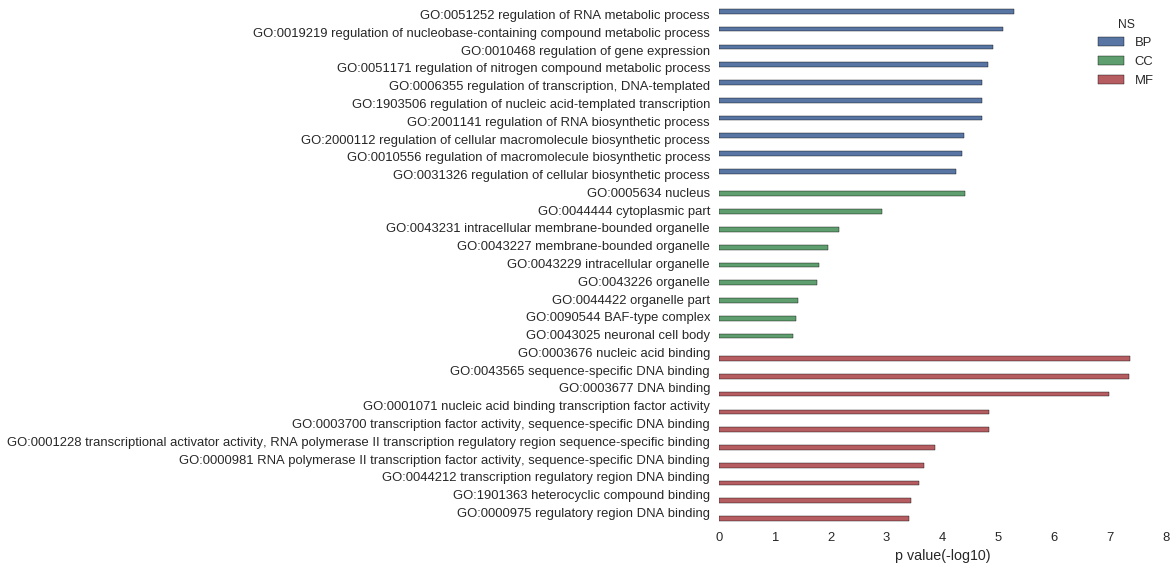
Gene Ontology
Download
| GO | P value |
|---|---|
| GO:0051252 | 5.18e-06 |
| GO:0019219 | 8.06e-06 |
| GO:0010468 | 1.25e-05 |
| GO:0051171 | 1.54e-05 |
| GO:0006355 | 1.91e-05 |
| GO:1903506 | 1.93e-05 |
| GO:2001141 | 1.96e-05 |
| GO:2000112 | 4.05e-05 |
| GO:0010556 | 4.38e-05 |
| GO:0031326 | 5.62e-05 |
| GO:0005634 | 3.96e-05 |
| GO:0044444 | 1.21e-03 |
| GO:0043231 | 7.15e-03 |
| GO:0043227 | 1.11e-02 |
| GO:0043229 | 1.63e-02 |
| GO:0043226 | 1.79e-02 |
| GO:0044422 | 3.85e-02 |
| GO:0090544 | 4.17e-02 |
| GO:0043025 | 4.80e-02 |
| GO:0003676 | 4.38e-08 |
| GO:0043565 | 4.48e-08 |
| GO:0003677 | 1.02e-07 |
| GO:0001071 | 1.46e-05 |
| GO:0003700 | 1.46e-05 |
| GO:0001228 | 1.38e-04 |
| GO:0000981 | 2.10e-04 |
| GO:0044212 | 2.62e-04 |
| GO:1901363 | 3.63e-04 |
| GO:0000975 | 3.91e-04 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01366
AGATACAAAGAAACACACACAAATACACACTAACACAGATACACACACAAATACACAAACACATATATAC
AGATACCCACACACACACACAAATAAACACACACACACACACACACACATATGTAATGTTGTAAATGTAC
TGCATTCACTGTACAACCCACCATTAATCTGATTTCTGATGAACCAAAGCTGTACACTGAGGAGTCTGCA
GAATGTGTGTTTTCTGGAGTGTGTGTGTGCGCGCATGTGTGTGTTTCTGGATGCAGAATAAAGGATCTGC
TTTAACAGACAGGCTGTGACCTCTGACCCCGGCCCGCCGCAGGGATGTGT
AGATACAAAGAAACACACACAAATACACACTAACACAGATACACACACAAATACACAAACACATATATAC
AGATACCCACACACACACACAAATAAACACACACACACACACACACACATATGTAATGTTGTAAATGTAC
TGCATTCACTGTACAACCCACCATTAATCTGATTTCTGATGAACCAAAGCTGTACACTGAGGAGTCTGCA
GAATGTGTGTTTTCTGGAGTGTGTGTGTGCGCGCATGTGTGTGTTTCTGGATGCAGAATAAAGGATCTGC
TTTAACAGACAGGCTGTGACCTCTGACCCCGGCCCGCCGCAGGGATGTGT