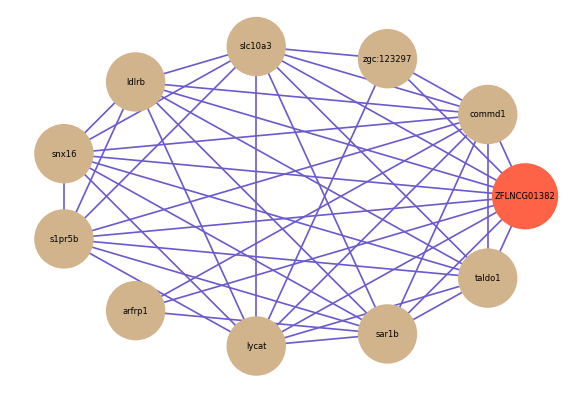
ZFLNCG01382
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 976.75 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 822.18 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 751.88 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 571.04 |
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 517.66 |
| ERR023143 | swim bladder | normal | 424.53 |
| ERR023146 | head kidney | normal | 336.19 |
| SRR516123 | skin | male and 3.5 year | 331.06 |
| SRR516121 | skin | male and 3.5 year | 210.68 |
| SRR594769 | blood | normal | 196.59 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
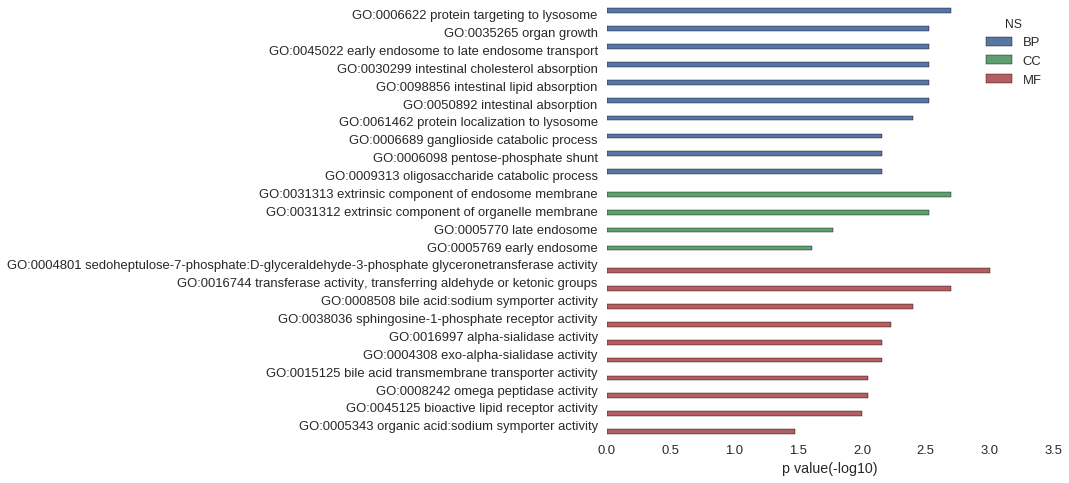
| GO | P value |
|---|---|
| GO:0006622 | 1.99e-03 |
| GO:0035265 | 2.98e-03 |
| GO:0045022 | 2.98e-03 |
| GO:0030299 | 2.98e-03 |
| GO:0098856 | 2.98e-03 |
| GO:0050892 | 2.98e-03 |
| GO:0061462 | 3.97e-03 |
| GO:0006689 | 6.94e-03 |
| GO:0006098 | 6.94e-03 |
| GO:0009313 | 6.94e-03 |
| GO:0031313 | 1.99e-03 |
| GO:0031312 | 2.98e-03 |
| GO:0005770 | 1.68e-02 |
| GO:0005769 | 2.46e-02 |
| GO:0004801 | 9.95e-04 |
| GO:0016744 | 1.99e-03 |
| GO:0008508 | 3.97e-03 |
| GO:0038036 | 5.96e-03 |
| GO:0016997 | 6.94e-03 |
| GO:0004308 | 6.94e-03 |
| GO:0015125 | 8.92e-03 |
| GO:0008242 | 8.92e-03 |
| GO:0045125 | 9.91e-03 |
| GO:0005343 | 3.33e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01382
TGGAAAAGGATCTCATGTTTATTCTGGTTCTCAAGGCATCAATTTCTCAGAGAAAATAACGACATCATTA
ATCAAGCACACGCTGCACGTCATCGGTTATTTGTTTTTAAAGCTCCACGTCTTGCAGCGACTCATTTGAG
CGTGTCTGTGTTTGGACTTTTATTCTGTAGTTTTGCTGACAGACTCGCACTTCCTGCTGCGCTAGAACAG
ACCAGACTGCTTTACATTCCCAAATAAAAGCAGTAAGATATTGCACACACACTCTCACTGCACTCACACA
CACACACACACACACACACACACTACACCCATACACAAACAGATCCACACACTAACACACATATATACAC
TAAACACATTCTATACTCACACATACTTGTGTACGCAAACAAACACACATACATACACTCACTCCCACAC
GCACTCACAATTACACCATCACACACATAATAAACAATACACACACACATACATTCCAAATATAAACTAC
ACTCATACACTTTACACACACATACATGCGCGCAAACACAT
TGGAAAAGGATCTCATGTTTATTCTGGTTCTCAAGGCATCAATTTCTCAGAGAAAATAACGACATCATTA
ATCAAGCACACGCTGCACGTCATCGGTTATTTGTTTTTAAAGCTCCACGTCTTGCAGCGACTCATTTGAG
CGTGTCTGTGTTTGGACTTTTATTCTGTAGTTTTGCTGACAGACTCGCACTTCCTGCTGCGCTAGAACAG
ACCAGACTGCTTTACATTCCCAAATAAAAGCAGTAAGATATTGCACACACACTCTCACTGCACTCACACA
CACACACACACACACACACACACTACACCCATACACAAACAGATCCACACACTAACACACATATATACAC
TAAACACATTCTATACTCACACATACTTGTGTACGCAAACAAACACACATACATACACTCACTCCCACAC
GCACTCACAATTACACCATCACACACATAATAAACAATACACACACACATACATTCCAAATATAAACTAC
ACTCATACACTTTACACACACATACATGCGCGCAAACACAT