ZFLNCG01506
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR891512 | blood | normal | 8.50 |
| SRR516124 | skin | male and 3.5 year | 8.07 |
| SRR516125 | skin | male and 3.5 year | 6.49 |
| SRR941753 | posterior pectoral fin | normal | 3.28 |
| SRR516129 | skin | male and 5 month | 2.75 |
| SRR516134 | skin | male and 5 month | 2.58 |
| SRR516131 | skin | male and 5 month | 1.72 |
| SRR516122 | skin | male and 3.5 year | 1.29 |
| SRR516123 | skin | male and 3.5 year | 1.16 |
| SRR516126 | skin | male and 5 month | 1.00 |
Express in tissues
Correlated coding gene
Download
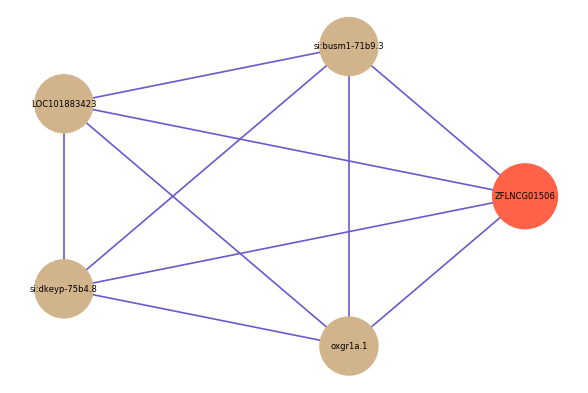
| gene | correlation coefficent |
|---|---|
| si:busm1-71b9.3 | 0.52 |
| LOC101883423 | 0.50 |
| si:dkeyp-75b4.8 | 0.50 |
| oxgr1a.1 | 0.50 |
Gene Ontology
Download
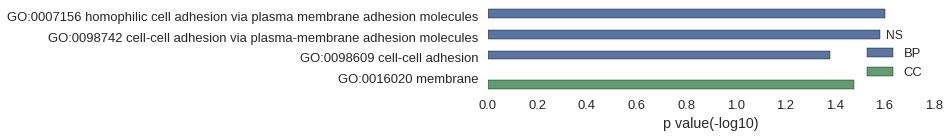
| GO | P value |
|---|---|
| GO:0007156 | 2.49e-02 |
| GO:0098742 | 2.62e-02 |
| GO:0098609 | 4.16e-02 |
| GO:0016020 | 3.34e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01506
AGTCCATAGCATCAGTGCCTTTGTCTTGCAGGAACTGCTGACACACTCCAGCCACATGAGGTCTAGCATT
GTCCTGCATTAGGAGGAACCCTGGGCCAACTACACTGGCATATAGTCTCACAAGGGGTCTGAGGATCTCA
TCTCGGTACCTAATGGCAGTCAGGCTACCTCTGGCAAGCCCATGGAGGGCTATGCGGCCCTCCAAAGAAA
TGCCACCCCAAAATGGGTCATGCTGCAGGATGTTGCAAGCAACACAATGTTCTCCACAGTGTCTCCAGAC
TCTGTCAAGTTTGTCAGTGTGAACCTGGTTTTATCTGTGAAGAGCACAGGGTGCCAGTGGCGAATTTGTC
TTGGTGTTCTTTGGCAAATGCCAAACGTCTTGCACGGTGTTGGGCTATAAGCACAACCCACACCTGTGGA
TGTCTGGCCCTCATACCACTCTCATGGTCTCTGTTTCTTACTGTTTGAGCAGACATATGCACATTTGGGG
CCTGCTGGAGGTCATTTTACTGTGCTCTGCCAGTGCTCCTCCTGTTCCTTCTTGCACAAAGACAAAGGTA
GCAGTCCTGCTGCTGGGTTGTTGCCCTACTACAGTCTCTTCCATGTCTTCTGATGTATTGGCCTGTCTCC
TTGTAACACCTTCATGCACTGGACAATACGCTGACAGTCACAGCAAACTTTCTTGCCACAGCTCGCATTG
ATGTGCCATCCTGGATGAGCTGCACTACCAGAGCCACTCGTGTGGGTTGTAGACAGGGCCGGCGCGTCCA
TAGAGGCAACCTAGGTGGCCACCTAAGGTGGCAAAATAGAGAGGGCGACGTCCGTGTCACCTCCCTCACC
ACCCACGTCCTCACAACACCCCCACCCCCATCCTCTATTTCATCCGTAACCCCCCGGAACTCCATCACAT
GCTACCACCGAAAGCACTGCCAGCATTCAAAAGTGACCAAAACATTAGCCAGAAAGCATAAGGACTGAGA
CGTGATCTGTGGTGAACACAGTGTCTTGCTAATTGCCTATAATTCCCTCTTTTTGTCTTTTCCATTTGCA
CAGCAGCATGTGAAATTGACTGTCCATCAGTTTTGCTTAATAAGTGGGTAGTTTGATTTCACAGAAGTGT
GATTGACTTTGAGCTACACTGTGTTGTTTGAGTGTTCTATTTATTTTTTCAAGCAGTGTATAATGATTCG
GT
AGTCCATAGCATCAGTGCCTTTGTCTTGCAGGAACTGCTGACACACTCCAGCCACATGAGGTCTAGCATT
GTCCTGCATTAGGAGGAACCCTGGGCCAACTACACTGGCATATAGTCTCACAAGGGGTCTGAGGATCTCA
TCTCGGTACCTAATGGCAGTCAGGCTACCTCTGGCAAGCCCATGGAGGGCTATGCGGCCCTCCAAAGAAA
TGCCACCCCAAAATGGGTCATGCTGCAGGATGTTGCAAGCAACACAATGTTCTCCACAGTGTCTCCAGAC
TCTGTCAAGTTTGTCAGTGTGAACCTGGTTTTATCTGTGAAGAGCACAGGGTGCCAGTGGCGAATTTGTC
TTGGTGTTCTTTGGCAAATGCCAAACGTCTTGCACGGTGTTGGGCTATAAGCACAACCCACACCTGTGGA
TGTCTGGCCCTCATACCACTCTCATGGTCTCTGTTTCTTACTGTTTGAGCAGACATATGCACATTTGGGG
CCTGCTGGAGGTCATTTTACTGTGCTCTGCCAGTGCTCCTCCTGTTCCTTCTTGCACAAAGACAAAGGTA
GCAGTCCTGCTGCTGGGTTGTTGCCCTACTACAGTCTCTTCCATGTCTTCTGATGTATTGGCCTGTCTCC
TTGTAACACCTTCATGCACTGGACAATACGCTGACAGTCACAGCAAACTTTCTTGCCACAGCTCGCATTG
ATGTGCCATCCTGGATGAGCTGCACTACCAGAGCCACTCGTGTGGGTTGTAGACAGGGCCGGCGCGTCCA
TAGAGGCAACCTAGGTGGCCACCTAAGGTGGCAAAATAGAGAGGGCGACGTCCGTGTCACCTCCCTCACC
ACCCACGTCCTCACAACACCCCCACCCCCATCCTCTATTTCATCCGTAACCCCCCGGAACTCCATCACAT
GCTACCACCGAAAGCACTGCCAGCATTCAAAAGTGACCAAAACATTAGCCAGAAAGCATAAGGACTGAGA
CGTGATCTGTGGTGAACACAGTGTCTTGCTAATTGCCTATAATTCCCTCTTTTTGTCTTTTCCATTTGCA
CAGCAGCATGTGAAATTGACTGTCCATCAGTTTTGCTTAATAAGTGGGTAGTTTGATTTCACAGAAGTGT
GATTGACTTTGAGCTACACTGTGTTGTTTGAGTGTTCTATTTATTTTTTCAAGCAGTGTATAATGATTCG
GT
