ZFLNCG01594
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 137.53 |
| SRR1648856 | brain | normal | 92.91 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 81.49 |
| SRR1048059 | pineal gland | light | 66.44 |
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 53.20 |
| SRR1648854 | brain | normal | 50.17 |
| SRR372802 | 5 dpf | normal | 35.35 |
| SRR1565811 | brain | normal | 19.21 |
| SRR594771 | endothelium | normal | 15.68 |
| SRR800045 | muscle | normal | 15.27 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| mmp16a | 0.53 |
| LOC101883507 | 0.51 |
| dixdc1a | 0.51 |
| zgc:100906 | 0.51 |
| magi2 | 0.51 |
| ptn | 0.51 |
| klf7b | 0.51 |
| amph | 0.51 |
| ahdc1 | 0.51 |
| gdi1 | 0.50 |
Gene Ontology
Download
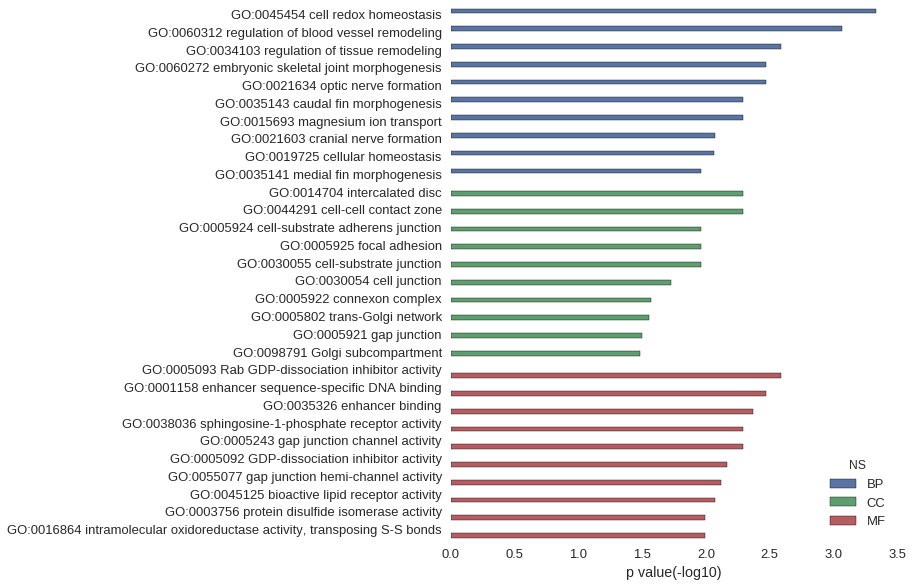
| GO | P value |
|---|---|
| GO:0045454 | 4.61e-04 |
| GO:0060312 | 8.53e-04 |
| GO:0034103 | 2.56e-03 |
| GO:0060272 | 3.41e-03 |
| GO:0021634 | 3.41e-03 |
| GO:0035143 | 5.11e-03 |
| GO:0015693 | 5.11e-03 |
| GO:0021603 | 8.50e-03 |
| GO:0019725 | 8.64e-03 |
| GO:0035141 | 1.10e-02 |
| GO:0014704 | 5.11e-03 |
| GO:0044291 | 5.11e-03 |
| GO:0005924 | 1.10e-02 |
| GO:0005925 | 1.10e-02 |
| GO:0030055 | 1.10e-02 |
| GO:0030054 | 1.89e-02 |
| GO:0005922 | 2.70e-02 |
| GO:0005802 | 2.78e-02 |
| GO:0005921 | 3.19e-02 |
| GO:0098791 | 3.28e-02 |
| GO:0005093 | 2.56e-03 |
| GO:0001158 | 3.41e-03 |
| GO:0035326 | 4.26e-03 |
| GO:0038036 | 5.11e-03 |
| GO:0005243 | 5.11e-03 |
| GO:0005092 | 6.80e-03 |
| GO:0055077 | 7.65e-03 |
| GO:0045125 | 8.50e-03 |
| GO:0003756 | 1.02e-02 |
| GO:0016864 | 1.02e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01594
TGACTTCTTGAGAAGTCCAAATTCTGGACCCTTCAAGGAGCTCATATAGTCGGTATTTGGAAAATCAACC
ACAGAGGAATGTTATAATGACACTGTTTAAAAAAAGGCGAGCAAGCCGAGGGGCCTTTTGTTCTGAGACA
GCCTGAAAAGAAAATTCATTTCCTGTCTGACAGATTACAGGATTACTAGGGGGAGAAAAAAAAACGCTAC
AAAGGGTACATGAGAAGTCAAAAGTGTCAGTTTTGCT
TGACTTCTTGAGAAGTCCAAATTCTGGACCCTTCAAGGAGCTCATATAGTCGGTATTTGGAAAATCAACC
ACAGAGGAATGTTATAATGACACTGTTTAAAAAAAGGCGAGCAAGCCGAGGGGCCTTTTGTTCTGAGACA
GCCTGAAAAGAAAATTCATTTCCTGTCTGACAGATTACAGGATTACTAGGGGGAGAAAAAAAAACGCTAC
AAAGGGTACATGAGAAGTCAAAAGTGTCAGTTTTGCT
