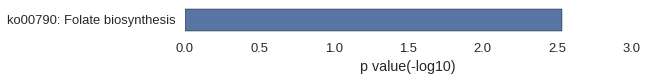
ZFLNCG01721
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR372787 | 2-4 cell | normal | 4.54 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 2.00 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 1.60 |
| SRR527835 | tail | normal | 1.57 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 1.39 |
| SRR748488 | sperm | normal | 0.65 |
| SRR748491 | germ ring | normal | 0.62 |
| SRR372793 | shield | normal | 0.36 |
| SRR748490 | 1 kcell | normal | 0.33 |
| SRR748489 | egg | normal | 0.24 |
Express in tissues
Correlated coding gene
Download
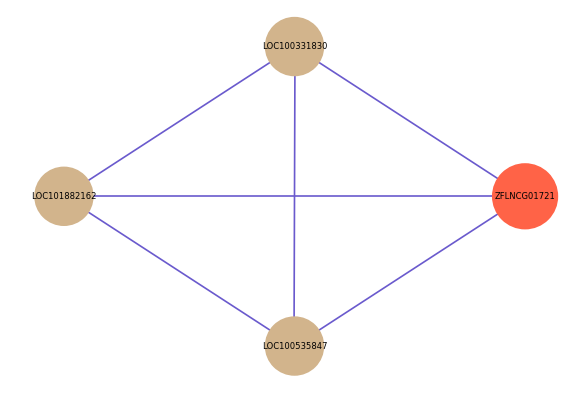
| gene | correlation coefficent |
|---|---|
| LOC100331830 | 0.55 |
| LOC101882162 | 0.55 |
| LOC100535847 | 0.50 |
Gene Ontology
Gene Ontology of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01721
CTAGTTATGTCTGGCTTCACTCCATGGTCTTTCTGTGGTTATCTCTGTAAATCTTGTAAATAAATATACT
TATTCAGATAATCCTGAGATCCCGTGCCTTCTTCGAAGTGAACCCAAGAGTTACATAATACCGGACCAAA
GAAAAAGTTTACGGCGTGTTTTCTCCTCGTTTTGTTTTCATAGTCTCCACTCCTCCTCCATGGTTTCTCC
CTCCATCGACTCCACCCTGGGCCATCCACCTGGCTGTGTCTTGGGTCCTCCTCCTGCCTCACCTTCCTCC
TAGAACTCCTCCCTGGCCTCCTCCTCCATTCCATCCCCTCCAACTCCCCTCCTGGATCCTGTGACTCCTC
ACTGGCCTCCTCCTGTTTCCATGCACTCTCTCCTGGTTCCTCCTCCACAGACTCCCCTTCACCCCCTCCT
TTCGTTTAGGTGCGGTGGTGTGCCTTTTAAAGGGGAGTGGGGAGGCGGGGAGGGGGTACTGTCACAACCT
GACTTCCTCGTGTGTTTATTTTTTTTCACCATGTGCTCCATTCTTCCAGTTCCAATCATTGTCATTACCC
TCATTGTGTATTTAAGTCCTTGTATTCAGTTCAGCCATTGTCTGCTATATTCTCACTAGTTATGTCTGTC
TTCACTCCATGTTCTTCCTGTGTTCATCTCCGTAAGTCTTGTAAATGAATAAATGTGCTTTGATAAACCT
GAGATTGCCTTCTTCAATGTGAACCCAGGCGTTACAGTCATAACGTATAATCGCG
CTAGTTATGTCTGGCTTCACTCCATGGTCTTTCTGTGGTTATCTCTGTAAATCTTGTAAATAAATATACT
TATTCAGATAATCCTGAGATCCCGTGCCTTCTTCGAAGTGAACCCAAGAGTTACATAATACCGGACCAAA
GAAAAAGTTTACGGCGTGTTTTCTCCTCGTTTTGTTTTCATAGTCTCCACTCCTCCTCCATGGTTTCTCC
CTCCATCGACTCCACCCTGGGCCATCCACCTGGCTGTGTCTTGGGTCCTCCTCCTGCCTCACCTTCCTCC
TAGAACTCCTCCCTGGCCTCCTCCTCCATTCCATCCCCTCCAACTCCCCTCCTGGATCCTGTGACTCCTC
ACTGGCCTCCTCCTGTTTCCATGCACTCTCTCCTGGTTCCTCCTCCACAGACTCCCCTTCACCCCCTCCT
TTCGTTTAGGTGCGGTGGTGTGCCTTTTAAAGGGGAGTGGGGAGGCGGGGAGGGGGTACTGTCACAACCT
GACTTCCTCGTGTGTTTATTTTTTTTCACCATGTGCTCCATTCTTCCAGTTCCAATCATTGTCATTACCC
TCATTGTGTATTTAAGTCCTTGTATTCAGTTCAGCCATTGTCTGCTATATTCTCACTAGTTATGTCTGTC
TTCACTCCATGTTCTTCCTGTGTTCATCTCCGTAAGTCTTGTAAATGAATAAATGTGCTTTGATAAACCT
GAGATTGCCTTCTTCAATGTGAACCCAGGCGTTACAGTCATAACGTATAATCGCG