ZFLNCG01865
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647683 | spleen | SVCV treatment | 19.19 |
| SRR1647680 | head kidney | SVCV treatment | 15.83 |
| SRR516132 | skin | male and 5 month | 10.30 |
| SRR1647682 | spleen | normal | 5.89 |
| SRR1647679 | head kidney | normal | 5.85 |
| SRR1562528 | eye | normal | 3.86 |
| SRR1647681 | head kidney | SVCV treatment | 3.70 |
| SRR1562531 | muscle | normal | 3.27 |
| SRR1647684 | spleen | SVCV treatment | 1.74 |
| SRR891512 | blood | normal | 1.66 |
Express in tissues
Correlated coding gene
Download
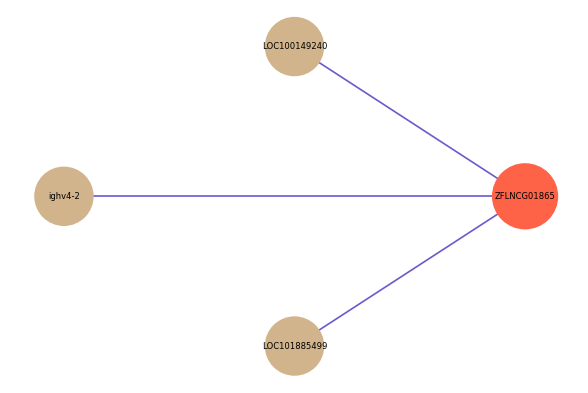
| gene | correlation coefficent |
|---|---|
| LOC100149240 | 0.57 |
| ighv4-2 | 0.52 |
| LOC101885499 | 0.51 |
Gene Ontology
Download
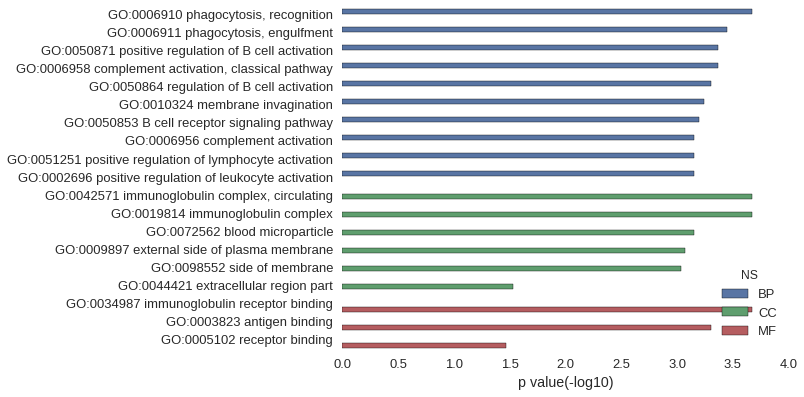
| GO | P value |
|---|---|
| GO:0006910 | 2.13e-04 |
| GO:0006911 | 3.55e-04 |
| GO:0050871 | 4.26e-04 |
| GO:0006958 | 4.26e-04 |
| GO:0050864 | 4.97e-04 |
| GO:0010324 | 5.69e-04 |
| GO:0050853 | 6.40e-04 |
| GO:0006956 | 7.11e-04 |
| GO:0051251 | 7.11e-04 |
| GO:0002696 | 7.11e-04 |
| GO:0042571 | 2.13e-04 |
| GO:0019814 | 2.13e-04 |
| GO:0072562 | 7.11e-04 |
| GO:0009897 | 8.53e-04 |
| GO:0098552 | 9.24e-04 |
| GO:0044421 | 2.95e-02 |
| GO:0034987 | 2.13e-04 |
| GO:0003823 | 4.97e-04 |
| GO:0005102 | 3.38e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01865
TTGTCTTGCACAGTAATACACGGCTGAATCTTCTGCTGTCAGGCCTGATAGTTTCAGATATGTCATACTC
TTGAATTATCTCTGGTGATTTCAGCTCTTCTTTTGAAGGATTCAGCATATTGGCCAGTGTATCCAATCCA
CACAAGAGGTTTTCCAGCTTGTTGTCTGATCCAGTTCATGTAATAGGAGCTAAACGTCAACCCAGATCCT
CTACAGGACAGAGTCAGAGTTTCTCCAGGCTTTCTCTGAACTGAGCTTTCAATCGACTCCATACTTTGAC
ATTTTAAACCTAAAAAAAAGACTGATTTAATTTGTAATATGTTTATTCTAGAACTGTCTATAGATCTTTA
TGTAAAACTCACACTGTAGACTGAATGAGAGCAGCAGTAAGCAGAGAGCATTCTTCAT
TTGTCTTGCACAGTAATACACGGCTGAATCTTCTGCTGTCAGGCCTGATAGTTTCAGATATGTCATACTC
TTGAATTATCTCTGGTGATTTCAGCTCTTCTTTTGAAGGATTCAGCATATTGGCCAGTGTATCCAATCCA
CACAAGAGGTTTTCCAGCTTGTTGTCTGATCCAGTTCATGTAATAGGAGCTAAACGTCAACCCAGATCCT
CTACAGGACAGAGTCAGAGTTTCTCCAGGCTTTCTCTGAACTGAGCTTTCAATCGACTCCATACTTTGAC
ATTTTAAACCTAAAAAAAAGACTGATTTAATTTGTAATATGTTTATTCTAGAACTGTCTATAGATCTTTA
TGTAAAACTCACACTGTAGACTGAATGAGAGCAGCAGTAAGCAGAGAGCATTCTTCAT
