ZFLNCG02110
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 77.31 |
| SRR516123 | skin | male and 3.5 year | 55.72 |
| SRR516122 | skin | male and 3.5 year | 37.52 |
| SRR516124 | skin | male and 3.5 year | 30.12 |
| SRR941749 | anterior pectoral fin | normal | 27.58 |
| ERR023143 | swim bladder | normal | 27.12 |
| SRR516121 | skin | male and 3.5 year | 25.93 |
| SRR516125 | skin | male and 3.5 year | 22.05 |
| SRR941753 | posterior pectoral fin | normal | 21.53 |
| ERR023146 | head kidney | normal | 21.34 |
Express in tissues
Correlated coding gene
Download
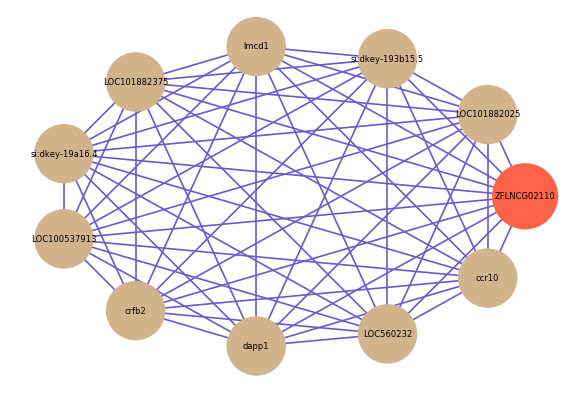
| gene | correlation coefficent |
|---|---|
| LOC101882025 | 0.71 |
| si:dkey-193b15.5 | 0.69 |
| lmcd1 | 0.68 |
| LOC101882375 | 0.68 |
| si:dkey-19a16.4 | 0.66 |
| LOC100537913 | 0.66 |
| crfb2 | 0.65 |
| dapp1 | 0.65 |
| LOC560232 | 0.64 |
| ccr10 | 0.64 |
Gene Ontology
Download
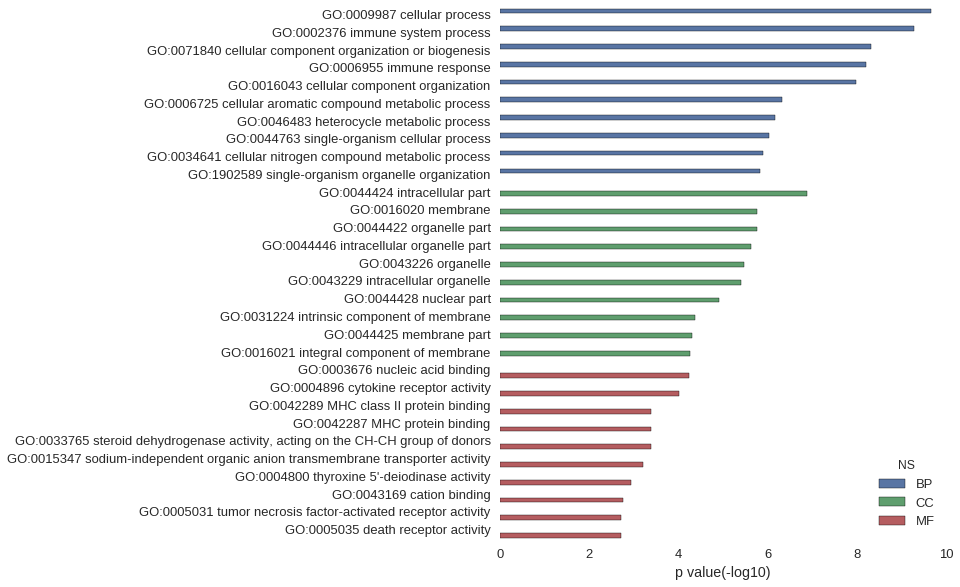
| GO | P value |
|---|---|
| GO:0009987 | 2.24e-10 |
| GO:0002376 | 5.14e-10 |
| GO:0071840 | 4.78e-09 |
| GO:0006955 | 6.19e-09 |
| GO:0016043 | 1.05e-08 |
| GO:0006725 | 4.77e-07 |
| GO:0046483 | 6.86e-07 |
| GO:0044763 | 9.35e-07 |
| GO:0034641 | 1.29e-06 |
| GO:1902589 | 1.50e-06 |
| GO:0044424 | 1.29e-07 |
| GO:0016020 | 1.69e-06 |
| GO:0044422 | 1.72e-06 |
| GO:0044446 | 2.34e-06 |
| GO:0043226 | 3.35e-06 |
| GO:0043229 | 4.03e-06 |
| GO:0044428 | 1.21e-05 |
| GO:0031224 | 4.15e-05 |
| GO:0044425 | 4.84e-05 |
| GO:0016021 | 5.35e-05 |
| GO:0003676 | 5.88e-05 |
| GO:0004896 | 9.89e-05 |
| GO:0042289 | 4.00e-04 |
| GO:0042287 | 4.00e-04 |
| GO:0033765 | 4.14e-04 |
| GO:0015347 | 6.12e-04 |
| GO:0004800 | 1.18e-03 |
| GO:0043169 | 1.78e-03 |
| GO:0005031 | 1.96e-03 |
| GO:0005035 | 1.96e-03 |
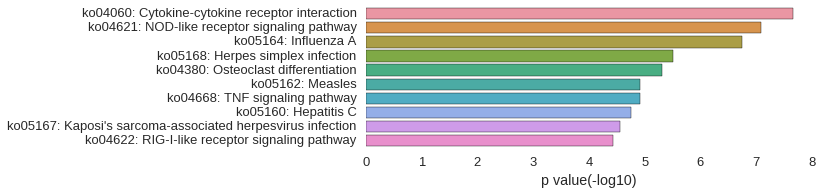
KEGG Pathway
Download
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT03102 | NONMMUT047084; | Direct blastn |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02110
AGCAGTACACACACACACACACACACACACACACGCACACACGCACACACACACATACACACACACACAC
ACACACACACACAAATAGAAAAATAAGAGGAAGTTTGACAATTCTCAGATTATTTAGATTATTAATATAC
GGCTGTCGACCAACATTATATCTGATAGTTGATGTGATTATTCAGTTTTATTATTATTTCATGTTAAAGA
CTTTTATGTTTATGTTCATGCACTCAACACCGTATTAAAGTCTGTTCAGAA
AGCAGTACACACACACACACACACACACACACACGCACACACGCACACACACACATACACACACACACAC
ACACACACACACAAATAGAAAAATAAGAGGAAGTTTGACAATTCTCAGATTATTTAGATTATTAATATAC
GGCTGTCGACCAACATTATATCTGATAGTTGATGTGATTATTCAGTTTTATTATTATTTCATGTTAAAGA
CTTTTATGTTTATGTTCATGCACTCAACACCGTATTAAAGTCTGTTCAGAA