ZFLNCG02215
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 4.03 |
| SRR748488 | sperm | normal | 3.15 |
| ERR023146 | head kidney | normal | 1.45 |
| SRR527835 | tail | normal | 0.44 |
| ERR145648 | skeletal muscle | 27 degree_C to 27 degree_C | 0.36 |
| SRR1647681 | head kidney | SVCV treatment | 0.33 |
| SRR748489 | egg | normal | 0.29 |
| SRR1562530 | liver | normal | 0.27 |
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 0.18 |
| SRR1028002 | head | normal | 0.16 |
Express in tissues
Correlated coding gene
Download
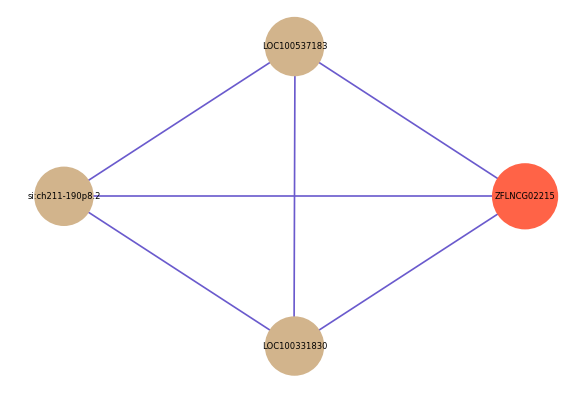
| gene | correlation coefficent |
|---|---|
| LOC100537183 | 0.53 |
| si:ch211-190p8.2 | 0.51 |
| LOC100331830 | 0.50 |
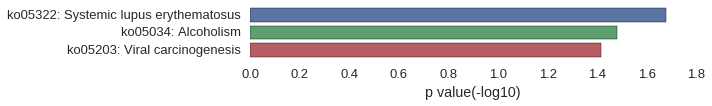
Gene Ontology
Download
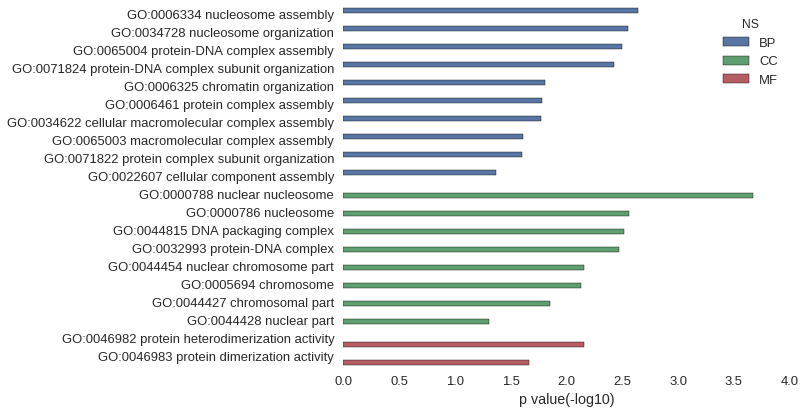
| GO | P value |
|---|---|
| GO:0006334 | 2.27e-03 |
| GO:0034728 | 2.84e-03 |
| GO:0065004 | 3.20e-03 |
| GO:0071824 | 3.77e-03 |
| GO:0006325 | 1.56e-02 |
| GO:0006461 | 1.67e-02 |
| GO:0034622 | 1.68e-02 |
| GO:0065003 | 2.47e-02 |
| GO:0071822 | 2.52e-02 |
| GO:0022607 | 4.26e-02 |
| GO:0000788 | 2.13e-04 |
| GO:0000786 | 2.77e-03 |
| GO:0044815 | 3.06e-03 |
| GO:0032993 | 3.41e-03 |
| GO:0044454 | 6.96e-03 |
| GO:0005694 | 7.46e-03 |
| GO:0044427 | 1.42e-02 |
| GO:0044428 | 4.99e-02 |
| GO:0046982 | 7.04e-03 |
| GO:0046983 | 2.19e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02215
TCACATTTAACCTTTTATTTTGCAAACAAAGACACCTGGCACATTTATACTACACAAAAACATTAACCAC
TCCAGGAGCCCTCCAACACTACATTTTCATGTATTAATTACAACTGATGTCCATCACCTCATTTAAAAAG
ACATGCAAAATGTGAAGTGCTAGTAAAGGCTCTATGAAGATGGTTTAAAGCACAGATTAACCAAACAATC
AGTCCAAGCTGATACCACCATCAGATGGATACAAGAGCAGCAGACAGAGGCTGCTCAGGAATAAGTCAAA
GGTGAATTAAGGTCAGCTGTTGAAGGACTCCATGCCAGTGTTACTCCTTCACCTCAGATCCAGAGACCTA
ATAATAAAAAGAAACATTGTTGTGGTTAAAGTCATTACCATAGTTAACATACAAGAAAATTGGGCAAATA
AACAATTAATTTCAATAAATCAATTGAAAATCGACATTAAAGAAAATGTTATAATTAAAGATTAAAAAGG
TGCAATTGTATAAAGGGAGCTGACTAAAATAACTTTAAATTACCCATTATTGAGACGAATGTGTTAGTCA
TCACTGCCTGGACATGTTAAAACTTAACATGTCAAAAGGAAGACAGATTTAAGAGCACTAATTATTTACA
AGGTTTCTTAGTGTTGTACTACTAAAAACCCATGAGCTTCTTCCAACTCATCCAGGAAGACTAAAATAAG
CACTTTAATGCACTTACAATACACACTTGGCAGATCTGTATACTACACACTCAAAACACTACATATTTGC
ACATCTTAAAACTGATGTCTATCACCTCATTTAAAGTCTACGAGACGTTAAAATGACATGCACAACTTCA
AATGCTAATAAAGTCCCAGTGAAAATGTTGAAAAACAGATTTAACCAAAGAATCAGTCAGAGCTGATACC
ACCATCAGATAGATCAAATAGCAGCAGACGGTTAGGGATAAAGTCTAGAGGTGAATAAAGTTAGGCTGTT
CAGGGACTTGCTACAATCACTCCTGCAGTTCTGATCCAGTGACCTAATTAAATAGAACAGTGAGAAAACA
G
TCACATTTAACCTTTTATTTTGCAAACAAAGACACCTGGCACATTTATACTACACAAAAACATTAACCAC
TCCAGGAGCCCTCCAACACTACATTTTCATGTATTAATTACAACTGATGTCCATCACCTCATTTAAAAAG
ACATGCAAAATGTGAAGTGCTAGTAAAGGCTCTATGAAGATGGTTTAAAGCACAGATTAACCAAACAATC
AGTCCAAGCTGATACCACCATCAGATGGATACAAGAGCAGCAGACAGAGGCTGCTCAGGAATAAGTCAAA
GGTGAATTAAGGTCAGCTGTTGAAGGACTCCATGCCAGTGTTACTCCTTCACCTCAGATCCAGAGACCTA
ATAATAAAAAGAAACATTGTTGTGGTTAAAGTCATTACCATAGTTAACATACAAGAAAATTGGGCAAATA
AACAATTAATTTCAATAAATCAATTGAAAATCGACATTAAAGAAAATGTTATAATTAAAGATTAAAAAGG
TGCAATTGTATAAAGGGAGCTGACTAAAATAACTTTAAATTACCCATTATTGAGACGAATGTGTTAGTCA
TCACTGCCTGGACATGTTAAAACTTAACATGTCAAAAGGAAGACAGATTTAAGAGCACTAATTATTTACA
AGGTTTCTTAGTGTTGTACTACTAAAAACCCATGAGCTTCTTCCAACTCATCCAGGAAGACTAAAATAAG
CACTTTAATGCACTTACAATACACACTTGGCAGATCTGTATACTACACACTCAAAACACTACATATTTGC
ACATCTTAAAACTGATGTCTATCACCTCATTTAAAGTCTACGAGACGTTAAAATGACATGCACAACTTCA
AATGCTAATAAAGTCCCAGTGAAAATGTTGAAAAACAGATTTAACCAAAGAATCAGTCAGAGCTGATACC
ACCATCAGATAGATCAAATAGCAGCAGACGGTTAGGGATAAAGTCTAGAGGTGAATAAAGTTAGGCTGTT
CAGGGACTTGCTACAATCACTCCTGCAGTTCTGATCCAGTGACCTAATTAAATAGAACAGTGAGAAAACA
G