ZFLNCG02281
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647683 | spleen | SVCV treatment | 31.08 |
| SRR1647684 | spleen | SVCV treatment | 29.94 |
| SRR1647682 | spleen | normal | 28.67 |
| SRR1562532 | spleen | normal | 25.31 |
| SRR1569491 | embryo | normal | 18.86 |
| SRR1648856 | brain | normal | 17.51 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 16.99 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 16.44 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 16.27 |
| SRR1648854 | brain | normal | 11.20 |
Express in tissues
Correlated coding gene
Download
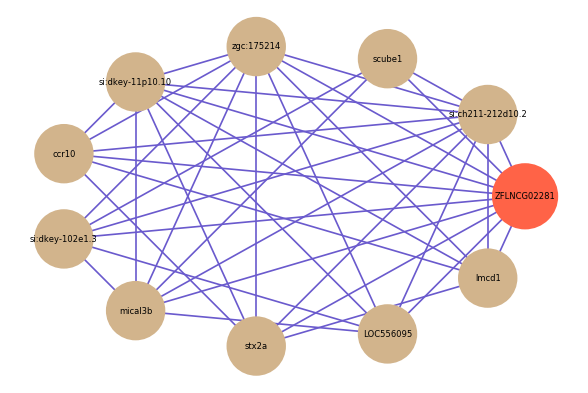
| gene | correlation coefficent |
|---|---|
| si:ch211-212d10.2 | 0.67 |
| scube1 | 0.65 |
| zgc:175214 | 0.65 |
| si:dkey-11p10.10 | 0.61 |
| ccr10 | 0.60 |
| si:dkey-102e1.3 | 0.59 |
| mical3b | 0.59 |
| stx2a | 0.58 |
| LOC556095 | 0.58 |
| lmcd1 | 0.58 |
Gene Ontology
Download
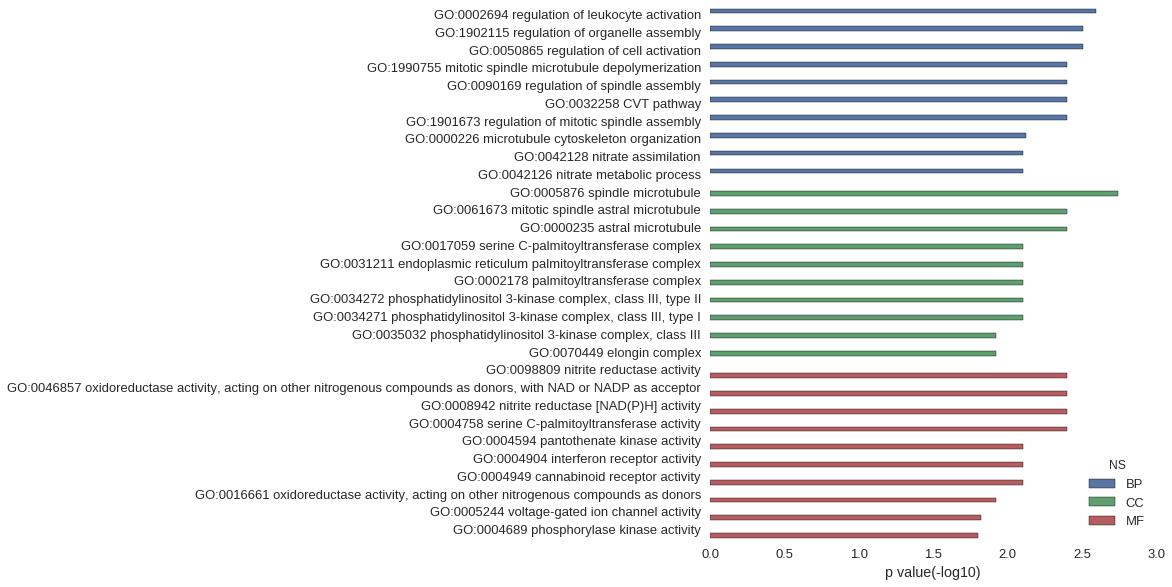
| GO | P value |
|---|---|
| GO:0002694 | 2.55e-03 |
| GO:1902115 | 3.11e-03 |
| GO:0050865 | 3.11e-03 |
| GO:1990755 | 3.98e-03 |
| GO:0090169 | 3.98e-03 |
| GO:0032258 | 3.98e-03 |
| GO:1901673 | 3.98e-03 |
| GO:0000226 | 7.55e-03 |
| GO:0042128 | 7.94e-03 |
| GO:0042126 | 7.94e-03 |
| GO:0005876 | 1.80e-03 |
| GO:0061673 | 3.98e-03 |
| GO:0000235 | 3.98e-03 |
| GO:0017059 | 7.94e-03 |
| GO:0031211 | 7.94e-03 |
| GO:0002178 | 7.94e-03 |
| GO:0034272 | 7.94e-03 |
| GO:0034271 | 7.94e-03 |
| GO:0035032 | 1.19e-02 |
| GO:0070449 | 1.19e-02 |
| GO:0098809 | 3.98e-03 |
| GO:0046857 | 3.98e-03 |
| GO:0008942 | 3.98e-03 |
| GO:0004758 | 3.98e-03 |
| GO:0004594 | 7.94e-03 |
| GO:0004904 | 7.94e-03 |
| GO:0004949 | 7.94e-03 |
| GO:0016661 | 1.19e-02 |
| GO:0005244 | 1.50e-02 |
| GO:0004689 | 1.58e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02281
CTTTATTCCTTAATATGCCAATGCATTTTAATAAAAGAGAAATGACAAAGTACAATAAAACAACCACAAC
ATGGGATTTGAATGATTTATTAATAAAGTGAGGTCTTAACAATAGTTTCCATTCTCTTATACACATCTTC
TGGGGTTGATTTCACACTGCTGAATTGTTCATTCTTCTGTACTGTGGTCATTTACCGTTGCGGGAATCAT
ACTATTCAACAGAGCCTTCCAGAGCAGTCATGTACATGTTAGACTGTCATCTTCCCCATCATAATGGCTC
ATTCTACTGTAAATATTAACTTAAGAATAAAAAAAACACTACCATTTGTACATCATTTACACATCTGAAT
TCATAAAGCTTACAATTCGCTCCAGCAGCAGTCTGATTCACTCTCTCCATCTGAGGGGCAAAGTACTTTG
AGATCACGTTGTGTGTGTGTGTAAATAAGTCGATTTCATGTCATACATTCTTCTTCTTTCTTCAAACAGT
CACGTGCCTTTCTTTAAATCAAATGAGACATTAAAATGTGATATAAAAATATTAATGACAACCTGGAGTT
ATCTCTGAAATGGCAAACAATATGGATTAGGTAATGCATCTGTGTTCATGCATGAATGTATGTTCATTTA
TGGTCGATGCATGAGCTAACGTGTTTCCCTGCCCATATGTGGATATTAGAAATACAAGCATTAATAAGTG
CTTATATAACAAAGCAGCTTTAGAATGATCACTCCGCCCCTTACGTCCAAAGCATATGCAAAACTATGCA
GAATACTGTAAGATGACAGAAAGGGTCATTTTCAACTTTATGGCACATCTACATGTGATACAATTATGAT
GTTTTTTTCCCCTAATATCTCACCCAAGTCTTGCAAAAAGATCCGTTTTTACAAAACAGACCAAGAATAT
TAAATAAAACTGTACCAGAACTACATGTAGAAACGTACAAACCTCTGATAGTATTTTTTGGCTTCCAACA
TGGTCATTCTGCTGATTTATGAGGTGAAACAATCTGAGCAAGTCAACATTCAAATACTTCAGATATTTCA
TCAGTTCAACAAACAGCCTCTTTTCTTCATCCACAGTTCAAAAACAAGAGCAAAATATGTGTCTTTGTGC
ACAGCACAAAGGTGATTTGTAATTAATTACTATGTAATAGAT
CTTTATTCCTTAATATGCCAATGCATTTTAATAAAAGAGAAATGACAAAGTACAATAAAACAACCACAAC
ATGGGATTTGAATGATTTATTAATAAAGTGAGGTCTTAACAATAGTTTCCATTCTCTTATACACATCTTC
TGGGGTTGATTTCACACTGCTGAATTGTTCATTCTTCTGTACTGTGGTCATTTACCGTTGCGGGAATCAT
ACTATTCAACAGAGCCTTCCAGAGCAGTCATGTACATGTTAGACTGTCATCTTCCCCATCATAATGGCTC
ATTCTACTGTAAATATTAACTTAAGAATAAAAAAAACACTACCATTTGTACATCATTTACACATCTGAAT
TCATAAAGCTTACAATTCGCTCCAGCAGCAGTCTGATTCACTCTCTCCATCTGAGGGGCAAAGTACTTTG
AGATCACGTTGTGTGTGTGTGTAAATAAGTCGATTTCATGTCATACATTCTTCTTCTTTCTTCAAACAGT
CACGTGCCTTTCTTTAAATCAAATGAGACATTAAAATGTGATATAAAAATATTAATGACAACCTGGAGTT
ATCTCTGAAATGGCAAACAATATGGATTAGGTAATGCATCTGTGTTCATGCATGAATGTATGTTCATTTA
TGGTCGATGCATGAGCTAACGTGTTTCCCTGCCCATATGTGGATATTAGAAATACAAGCATTAATAAGTG
CTTATATAACAAAGCAGCTTTAGAATGATCACTCCGCCCCTTACGTCCAAAGCATATGCAAAACTATGCA
GAATACTGTAAGATGACAGAAAGGGTCATTTTCAACTTTATGGCACATCTACATGTGATACAATTATGAT
GTTTTTTTCCCCTAATATCTCACCCAAGTCTTGCAAAAAGATCCGTTTTTACAAAACAGACCAAGAATAT
TAAATAAAACTGTACCAGAACTACATGTAGAAACGTACAAACCTCTGATAGTATTTTTTGGCTTCCAACA
TGGTCATTCTGCTGATTTATGAGGTGAAACAATCTGAGCAAGTCAACATTCAAATACTTCAGATATTTCA
TCAGTTCAACAAACAGCCTCTTTTCTTCATCCACAGTTCAAAAACAAGAGCAAAATATGTGTCTTTGTGC
ACAGCACAAAGGTGATTTGTAATTAATTACTATGTAATAGAT