ZFLNCG02364
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1004789 | larvae | impdh2 morpholino | 65.74 |
| SRR941753 | posterior pectoral fin | normal | 59.27 |
| SRR1104058 | heart | normal | 52.63 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 51.19 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 48.24 |
| SRR797908 | embryo | normal | 46.97 |
| SRR941749 | anterior pectoral fin | normal | 45.79 |
| SRR1523214 | embryo | glut12 morpholino | 44.36 |
| SRR1523211 | embryo | control morpholino | 43.01 |
| SRR1035984 | 13 hpf | normal | 42.93 |
Express in tissues
Correlated coding gene
Download
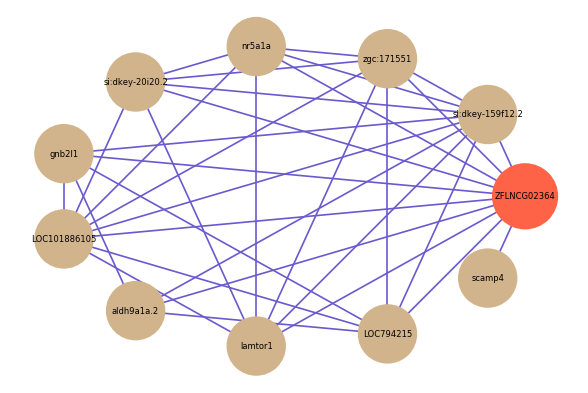
| gene | correlation coefficent |
|---|---|
| si:dkey-159f12.2 | 0.63 |
| zgc:171551 | 0.62 |
| nr5a1a | 0.62 |
| si:dkey-20i20.2 | 0.60 |
| gnb2l1 | 0.58 |
| LOC101886105 | 0.58 |
| aldh9a1a.2 | 0.57 |
| lamtor1 | 0.57 |
| LOC794215 | 0.56 |
| scamp4 | 0.56 |
Gene Ontology
Download
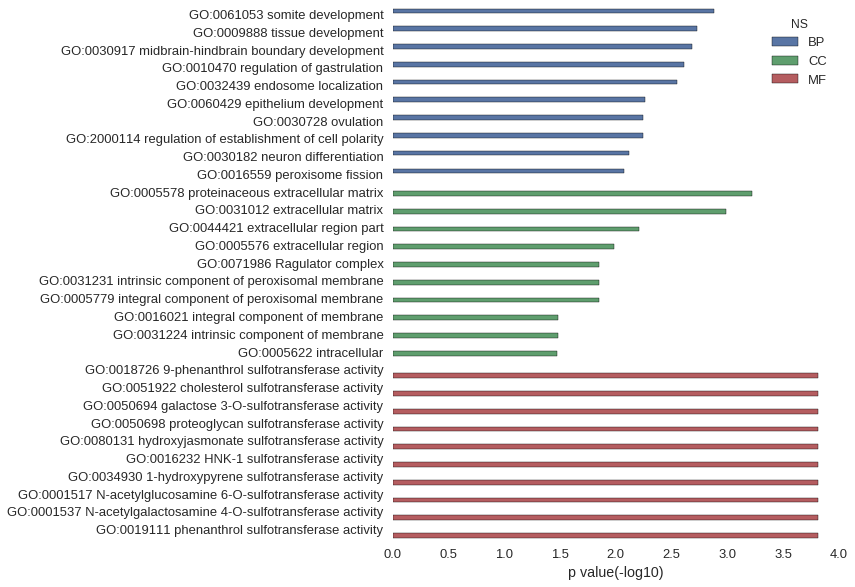
| GO | P value |
|---|---|
| GO:0061053 | 1.31e-03 |
| GO:0009888 | 1.86e-03 |
| GO:0030917 | 2.09e-03 |
| GO:0010470 | 2.45e-03 |
| GO:0032439 | 2.84e-03 |
| GO:0060429 | 5.44e-03 |
| GO:0030728 | 5.68e-03 |
| GO:2000114 | 5.68e-03 |
| GO:0030182 | 7.57e-03 |
| GO:0016559 | 8.50e-03 |
| GO:0005578 | 5.99e-04 |
| GO:0031012 | 1.02e-03 |
| GO:0044421 | 6.13e-03 |
| GO:0005576 | 1.04e-02 |
| GO:0071986 | 1.41e-02 |
| GO:0031231 | 1.41e-02 |
| GO:0005779 | 1.41e-02 |
| GO:0016021 | 3.27e-02 |
| GO:0031224 | 3.28e-02 |
| GO:0005622 | 3.33e-02 |
| GO:0018726 | 1.55e-04 |
| GO:0051922 | 1.55e-04 |
| GO:0050694 | 1.55e-04 |
| GO:0050698 | 1.55e-04 |
| GO:0080131 | 1.55e-04 |
| GO:0016232 | 1.55e-04 |
| GO:0034930 | 1.55e-04 |
| GO:0001517 | 1.55e-04 |
| GO:0001537 | 1.55e-04 |
| GO:0019111 | 1.55e-04 |
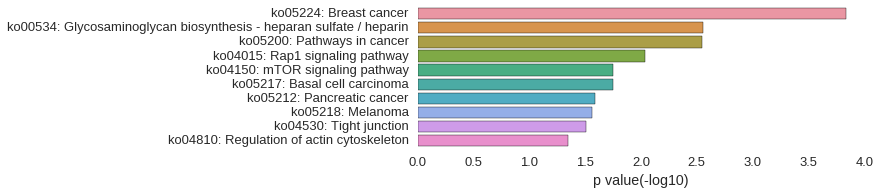
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02364
ACGAAGTGAGGTATTTTATTCCCGTTGCTGTTAGAAAATATTCACAACTGGTGTCATTTGCTCCCGTACA
GCTGTCAGAAAGTAAACCAGCGGTCTTTGTAAAGGACGCGGAGGGGCTCAGTATCCAAATATCGCGAAGC
AATGGGTCAGCATAGCGGAGACGCAGAAAGCCAGTTTGAATCCCAGCTGCGATGAGTAAACAT
ACGAAGTGAGGTATTTTATTCCCGTTGCTGTTAGAAAATATTCACAACTGGTGTCATTTGCTCCCGTACA
GCTGTCAGAAAGTAAACCAGCGGTCTTTGTAAAGGACGCGGAGGGGCTCAGTATCCAAATATCGCGAAGC
AATGGGTCAGCATAGCGGAGACGCAGAAAGCCAGTTTGAATCCCAGCTGCGATGAGTAAACAT