ZFLNCG02373
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035237 | liver | transgenic UHRF1 | 31.55 |
| SRR891495 | heart | normal | 9.31 |
| SRR1647681 | head kidney | SVCV treatment | 9.22 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 8.57 |
| SRR891504 | liver | normal | 8.55 |
| SRR891512 | blood | normal | 8.21 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 7.71 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 7.50 |
| SRR1647679 | head kidney | normal | 7.36 |
| SRR527835 | tail | normal | 7.33 |
Express in tissues
Correlated coding gene
Download
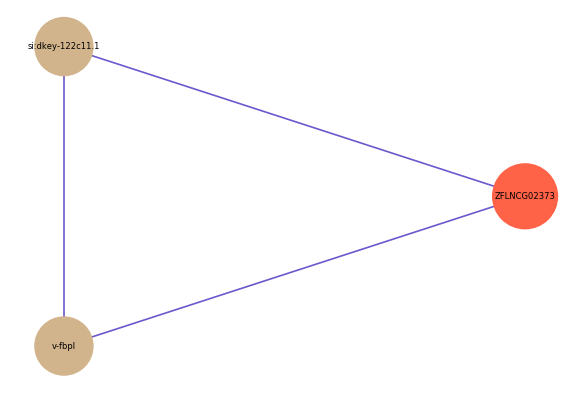
| gene | correlation coefficent |
|---|---|
| si:dkey-122c11.1 | 0.55 |
| v-fbpl | 0.51 |
Gene Ontology
Gene Ontology of this lncRNA gene hava does not exist!
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT03700 | ENST00000548437; ENST00000552999; ENST00000549403; NONHSAT027803; NONHSAT027802; NONHSAT027804; | Collinearity with conserved coding gene |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02373
AATACAGCTCCTCATATTTGACGTTTTTTAGTCTCTAAACACTTTTAATCAGTGTCATTGAGTAACCTAT
GAAAGATTAGTCTTCATAATCTCTCCGAGACACACAGTAGCGTAGCAGGACGCCTCCAGGTCAAAGTTTA
GTGACAGACAGTGTGTGTTAGACTCGTCTGATGAACTCGTCTGCAGGCTATATGTGATGTTCTGTGGCTT
AGCGAGCAGGGGTCGATAACATCCAGGGACGTTGAGTTTAAGGGGAGTCACTCTGAATCGGCAGGACTCC
AAACCGTCTTGAGCCAATCGGTCTTGATACCATTGACCCAAAAGGTTTTCTGGATACTTGACACTATTTC
CTGGCATAGGAAGAATCACCTGGATTAAAAACCACAGAGGCACAATAAGAGACTGATGGGTAAGAGTTAC
TGTAGCAGACATACAGAGCAATGAAAGACATCTGAAACAGTACCTGATCTAATGAGAAAACCTCATTCTT
CTCCTCATCTGAAGTCACAACATGGATCTACAGCACAGAAAACAAGTGAAAATGAATGGAAAACCACAAA
TAAGAACAAGAAACAATAATACAAATGTACAATCATTCAACATACCTGCATTTATGTACACACACTCTTC
TTTAATAAAATATTTGAAAGATTTATGCATGTTTAATTTATATATACATTTTTAAAATACTTCTTTACAA
AGTAGACTCAAATAATCAAACAAATGTTTCTTAACGTTGTATTTCAGGGTAAATGTTAGGATATTATTAG
ATTGAAATTTACTCTGAATAAAGTGCGTGTTTGTTTTATTCTTATGATTTATTTCACCCAATTCAATCTA
AACACAAACCGTTTCTGGCTATTATTTAGCAATAACTAAATCAAGACCCTTACCTGTGGAGCACTGAGCT
CCTCTGTGGCATCTTTGAGCCCTTTTACAGGCTCAGCCCAAACCAGATCGCCCTGAACAGGCTTAAAGCC
CAGTTTTTGCAGCCGGTATTTGGCTGCTTGGTTCCACACTCTGCTGCAGTAGGAGTGAAGGTAGAAGACC
CTCATGCTGTGGGGTAAACTGAGCCAGCCACGAGTGCAACCCTCCTGTCCACTGCCGTACCGGTGGAGCG
CCCGCAGCATGA
AATACAGCTCCTCATATTTGACGTTTTTTAGTCTCTAAACACTTTTAATCAGTGTCATTGAGTAACCTAT
GAAAGATTAGTCTTCATAATCTCTCCGAGACACACAGTAGCGTAGCAGGACGCCTCCAGGTCAAAGTTTA
GTGACAGACAGTGTGTGTTAGACTCGTCTGATGAACTCGTCTGCAGGCTATATGTGATGTTCTGTGGCTT
AGCGAGCAGGGGTCGATAACATCCAGGGACGTTGAGTTTAAGGGGAGTCACTCTGAATCGGCAGGACTCC
AAACCGTCTTGAGCCAATCGGTCTTGATACCATTGACCCAAAAGGTTTTCTGGATACTTGACACTATTTC
CTGGCATAGGAAGAATCACCTGGATTAAAAACCACAGAGGCACAATAAGAGACTGATGGGTAAGAGTTAC
TGTAGCAGACATACAGAGCAATGAAAGACATCTGAAACAGTACCTGATCTAATGAGAAAACCTCATTCTT
CTCCTCATCTGAAGTCACAACATGGATCTACAGCACAGAAAACAAGTGAAAATGAATGGAAAACCACAAA
TAAGAACAAGAAACAATAATACAAATGTACAATCATTCAACATACCTGCATTTATGTACACACACTCTTC
TTTAATAAAATATTTGAAAGATTTATGCATGTTTAATTTATATATACATTTTTAAAATACTTCTTTACAA
AGTAGACTCAAATAATCAAACAAATGTTTCTTAACGTTGTATTTCAGGGTAAATGTTAGGATATTATTAG
ATTGAAATTTACTCTGAATAAAGTGCGTGTTTGTTTTATTCTTATGATTTATTTCACCCAATTCAATCTA
AACACAAACCGTTTCTGGCTATTATTTAGCAATAACTAAATCAAGACCCTTACCTGTGGAGCACTGAGCT
CCTCTGTGGCATCTTTGAGCCCTTTTACAGGCTCAGCCCAAACCAGATCGCCCTGAACAGGCTTAAAGCC
CAGTTTTTGCAGCCGGTATTTGGCTGCTTGGTTCCACACTCTGCTGCAGTAGGAGTGAAGGTAGAAGACC
CTCATGCTGTGGGGTAAACTGAGCCAGCCACGAGTGCAACCCTCCTGTCCACTGCCGTACCGGTGGAGCG
CCCGCAGCATGA
