ZFLNCG02423
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR658547 | 6 somite | Gata5/6 morphant | 12.22 |
| SRR658539 | bud | Gata5/6 morphant | 11.61 |
| SRR658543 | 6 somite | Gata5 morphan | 11.22 |
| SRR658535 | bud | Gata5 morphant | 11.00 |
| SRR658541 | 6 somite | normal | 10.73 |
| SRR1569491 | embryo | normal | 10.01 |
| SRR1004785 | larvae | normal | 9.36 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 9.34 |
| SRR658537 | bud | Gata6 morphant | 9.02 |
| SRR658533 | bud | normal | 8.32 |
Express in tissues
Correlated coding gene
Download
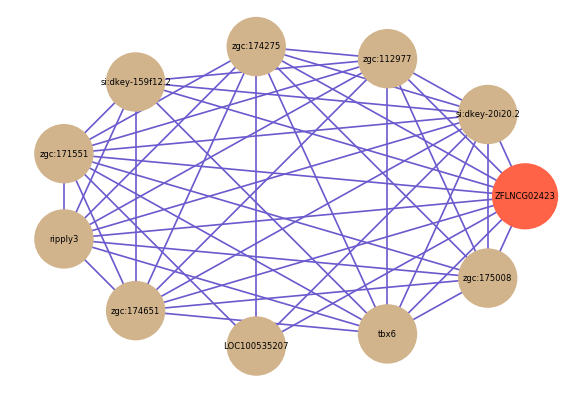
| gene | correlation coefficent |
|---|---|
| si:dkey-20i20.2 | 0.57 |
| zgc:112977 | 0.54 |
| zgc:174275 | 0.53 |
| si:dkey-159f12.2 | 0.53 |
| zgc:171551 | 0.52 |
| ripply3 | 0.52 |
| zgc:174651 | 0.52 |
| LOC100535207 | 0.51 |
| tbx6 | 0.51 |
| zgc:175008 | 0.51 |
Gene Ontology
Download
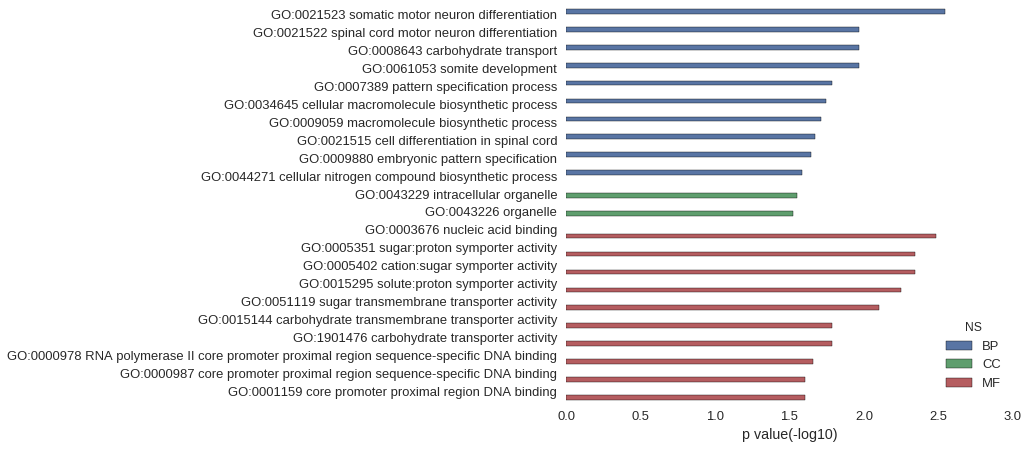
| GO | P value |
|---|---|
| GO:0021523 | 2.84e-03 |
| GO:0021522 | 1.08e-02 |
| GO:0008643 | 1.08e-02 |
| GO:0061053 | 1.08e-02 |
| GO:0007389 | 1.63e-02 |
| GO:0034645 | 1.81e-02 |
| GO:0009059 | 1.95e-02 |
| GO:0021515 | 2.14e-02 |
| GO:0009880 | 2.25e-02 |
| GO:0044271 | 2.61e-02 |
| GO:0043229 | 2.80e-02 |
| GO:0043226 | 3.01e-02 |
| GO:0003676 | 3.26e-03 |
| GO:0005351 | 4.54e-03 |
| GO:0005402 | 4.54e-03 |
| GO:0015295 | 5.67e-03 |
| GO:0051119 | 7.93e-03 |
| GO:0015144 | 1.64e-02 |
| GO:1901476 | 1.64e-02 |
| GO:0000978 | 2.20e-02 |
| GO:0000987 | 2.47e-02 |
| GO:0001159 | 2.47e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02423
AACTCCTCCACAATAAATCACCTTACGACATTAACATGAGCAGTTTTGCTAGCTAAATCCATGACATAAT
ACAGGAGTGAATTAAACTGTCTGTAAATGATGTGTTTTGCCTGCAGTTATCCACTCGAGAAAAGACCCGC
ACATGTTGACAAGGCGAGACATTTCAGGTAAATTTTTCTGAAGATCAGTTTGTTTATGATATTGTTAAAA
TGTGACTGACGATGGTCTTTTCTTACAGATGGTCAAGGGCAAATGAGAATCTTTTTGCTACCACAGGCTA
CCCAGGAAAAATCAACAGTCAGTTACTGGTGCATCATCTTGGCCATCCACAGGTTTATTCATTTATATTT
ATATACAGAACATGCTTGACGGTTTTAAATAGTACGTTTGCATCTGCCTCAGTTCTCAAGGTATGTAATT
GTCATTGCAGCCTGTCATGCTTGGTTCGGCATCTGTGGGGGCTGGACTGAGCTGGCACAGGACCCTGCCT
CTTTGTGTTATTGGTGGGGATCGAAAGTTGTTCTTCTGGATGACGGAGATGTAAATTGGTTCGAAGTCAA
CTGGAGAGTCTTTCTGTTTTCCTTTTTCAGTAAATAAAGTCATATTTTTCATGC
AACTCCTCCACAATAAATCACCTTACGACATTAACATGAGCAGTTTTGCTAGCTAAATCCATGACATAAT
ACAGGAGTGAATTAAACTGTCTGTAAATGATGTGTTTTGCCTGCAGTTATCCACTCGAGAAAAGACCCGC
ACATGTTGACAAGGCGAGACATTTCAGGTAAATTTTTCTGAAGATCAGTTTGTTTATGATATTGTTAAAA
TGTGACTGACGATGGTCTTTTCTTACAGATGGTCAAGGGCAAATGAGAATCTTTTTGCTACCACAGGCTA
CCCAGGAAAAATCAACAGTCAGTTACTGGTGCATCATCTTGGCCATCCACAGGTTTATTCATTTATATTT
ATATACAGAACATGCTTGACGGTTTTAAATAGTACGTTTGCATCTGCCTCAGTTCTCAAGGTATGTAATT
GTCATTGCAGCCTGTCATGCTTGGTTCGGCATCTGTGGGGGCTGGACTGAGCTGGCACAGGACCCTGCCT
CTTTGTGTTATTGGTGGGGATCGAAAGTTGTTCTTCTGGATGACGGAGATGTAAATTGGTTCGAAGTCAA
CTGGAGAGTCTTTCTGTTTTCCTTTTTCAGTAAATAAAGTCATATTTTTCATGC
