ZFLNCG02743
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR372791 | dome | normal | 1.22 |
| SRR748491 | germ ring | normal | 0.60 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 0.36 |
| SRR1021215 | sphere stage | Mzeomesa | 0.35 |
| SRR372793 | shield | normal | 0.30 |
| SRR797914 | embryo | pbx2/4-MO | 0.29 |
| SRR372796 | bud | normal | 0.24 |
| SRR658533 | bud | normal | 0.23 |
| SRR800043 | sphere stage | normal | 0.22 |
| SRR658543 | 6 somite | Gata5 morphan | 0.20 |
Express in tissues
Correlated coding gene
Download
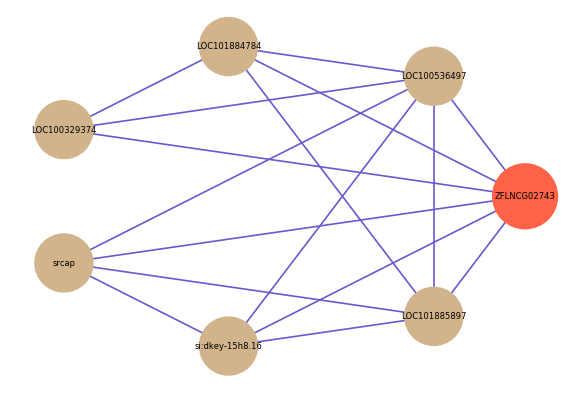
| gene | correlation coefficent |
|---|---|
| LOC100536497 | 0.54 |
| LOC101884784 | 0.52 |
| LOC100329374 | 0.51 |
| srcap | 0.51 |
| si:dkey-15h8.16 | 0.51 |
| LOC101885897 | 0.50 |
Gene Ontology
Download
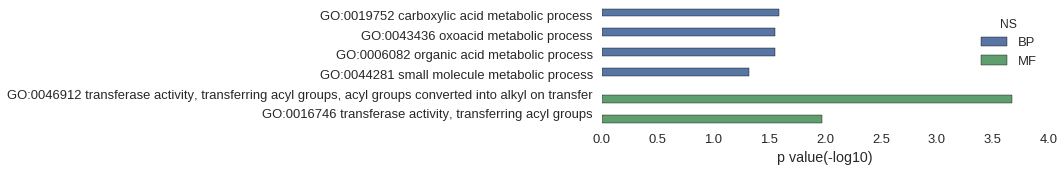
| GO | P value |
|---|---|
| GO:0019752 | 2.58e-02 |
| GO:0043436 | 2.79e-02 |
| GO:0006082 | 2.81e-02 |
| GO:0044281 | 4.81e-02 |
| GO:0046912 | 2.13e-04 |
| GO:0016746 | 1.07e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02743
TGGATTGTTGGTGATTGTATGGGTTTTGGCCGGATTGGGTAACAAAATATGAACAAACGAAAATAAAGAC
TTGCTTAAAAAAGATTTAAGACCTACAACACAATATTTCAGTAAATTTAGGACTTTTGAAGACCTAAAAT
TTTGATTTAGGAATGTACGACATTTTAGGACCCCACGGATACCCTGTTTAAACACAATAGCGCTGGTAAT
TAGTTATTATAATATTGATGTAATTCTAACTCAGATACTATCTGTGAGAACTAGTGACAACTACTAGAAA
CAACTTTAAACCCATAAAGAAGTTGATTAAAAAAGGGTAATTGTGATCAACGTGACATATGCAAATGGAA
AGTACTAAGCCAACACTATCACTGTAATAGCAGATATCTTCTATGAGAATAATGTAGGTCAAATTTTGTC
AGCTCAGTTAAACTTTTTTTTAATTTATTGTTTTTATTTATTTATTTATATATAGTGCCAGTGCGCAGGG
TACAGTCGGAGAACAAGAAAAGAAGGTTAAAAAGGTAGAGAAAATAGGAGACTAATAATACATAAAATAA
TGCCAAAAGACATGAGAACAAGTAAGAAAGAAACTCATTCCTGTCTTTAAATGACTGCTTATGTGCATTT
AGTAGAACTAGGCAGCACACTCAGGGCTGCAAGATGGATTTAATTTATTATTCTTATTATTAAAATATAT
CTGAATGAGGATCAAATGACTGAGTTGGTCTTTAAGAATGAAAACTAACCTGTTTGTTCCTGCGGATCTT
CCTGTTTGACTGTGAATGTTTCTTCAATCTCCACATCTTCACTCTCCACTTTAATAAACGCCATCTTTAT
AACAGTGTGGAGATCAGTCGCTTCAGCATGAATTTTTCTCTGTGTTTGGACACTTTATCCTGTTTAAAAC
TAATAAAACAATAAACAGAAACAAAATAACTTCTCTTTAACACTTGCTTCAATGGTTTCTTTATATGAGA
GTAATTAACAAAACGTGTCTTTACAACACTTAACTTATTACACACTTTCCTGTTACTTTATTCAAACAAT
AGCCATTAGTTACTGAGAATAAAATAGTACTACGTTGTATAAAGGGTTAAAATAATATTTCCAACAGCAG
TAACATAATATAGCATCGTATAAAAAACTTAAAACTTTAGAATTTTAACTTTATATCAGTTTATATCTGT
AAACGTTTTCAAACAAACCTTTCTCCAGTTGAATCACAGCGACGAAGCGGCGCTGCCTGATGACGTCACG
CG
TGGATTGTTGGTGATTGTATGGGTTTTGGCCGGATTGGGTAACAAAATATGAACAAACGAAAATAAAGAC
TTGCTTAAAAAAGATTTAAGACCTACAACACAATATTTCAGTAAATTTAGGACTTTTGAAGACCTAAAAT
TTTGATTTAGGAATGTACGACATTTTAGGACCCCACGGATACCCTGTTTAAACACAATAGCGCTGGTAAT
TAGTTATTATAATATTGATGTAATTCTAACTCAGATACTATCTGTGAGAACTAGTGACAACTACTAGAAA
CAACTTTAAACCCATAAAGAAGTTGATTAAAAAAGGGTAATTGTGATCAACGTGACATATGCAAATGGAA
AGTACTAAGCCAACACTATCACTGTAATAGCAGATATCTTCTATGAGAATAATGTAGGTCAAATTTTGTC
AGCTCAGTTAAACTTTTTTTTAATTTATTGTTTTTATTTATTTATTTATATATAGTGCCAGTGCGCAGGG
TACAGTCGGAGAACAAGAAAAGAAGGTTAAAAAGGTAGAGAAAATAGGAGACTAATAATACATAAAATAA
TGCCAAAAGACATGAGAACAAGTAAGAAAGAAACTCATTCCTGTCTTTAAATGACTGCTTATGTGCATTT
AGTAGAACTAGGCAGCACACTCAGGGCTGCAAGATGGATTTAATTTATTATTCTTATTATTAAAATATAT
CTGAATGAGGATCAAATGACTGAGTTGGTCTTTAAGAATGAAAACTAACCTGTTTGTTCCTGCGGATCTT
CCTGTTTGACTGTGAATGTTTCTTCAATCTCCACATCTTCACTCTCCACTTTAATAAACGCCATCTTTAT
AACAGTGTGGAGATCAGTCGCTTCAGCATGAATTTTTCTCTGTGTTTGGACACTTTATCCTGTTTAAAAC
TAATAAAACAATAAACAGAAACAAAATAACTTCTCTTTAACACTTGCTTCAATGGTTTCTTTATATGAGA
GTAATTAACAAAACGTGTCTTTACAACACTTAACTTATTACACACTTTCCTGTTACTTTATTCAAACAAT
AGCCATTAGTTACTGAGAATAAAATAGTACTACGTTGTATAAAGGGTTAAAATAATATTTCCAACAGCAG
TAACATAATATAGCATCGTATAAAAAACTTAAAACTTTAGAATTTTAACTTTATATCAGTTTATATCTGT
AAACGTTTTCAAACAAACCTTTCTCCAGTTGAATCACAGCGACGAAGCGGCGCTGCCTGATGACGTCACG
CG
