ZFLNCG02761
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 3.31 |
| SRR658539 | bud | Gata5/6 morphant | 1.40 |
| SRR658537 | bud | Gata6 morphant | 1.39 |
| SRR658547 | 6 somite | Gata5/6 morphant | 1.33 |
| SRR1021215 | sphere stage | Mzeomesa | 1.01 |
| SRR658533 | bud | normal | 0.87 |
| SRR800043 | sphere stage | normal | 0.85 |
| SRR1021213 | sphere stage | normal | 0.70 |
| SRR800049 | sphere stage | control treatment | 0.65 |
| SRR700537 | heart | Gata4 morpholino | 0.64 |
Express in tissues
Correlated coding gene
Download
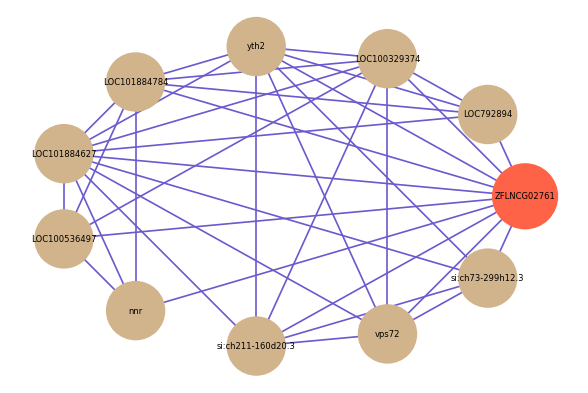
| gene | correlation coefficent |
|---|---|
| LOC792894 | 0.55 |
| LOC100536497 | 0.54 |
| si:ch211-160d20.3 | 0.53 |
| nnr | 0.52 |
| LOC100329374 | 0.52 |
| yth2 | 0.52 |
| LOC101884784 | 0.52 |
| LOC101884627 | 0.52 |
| vps72 | 0.52 |
| si:ch73-299h12.3 | 0.52 |
Gene Ontology
Download
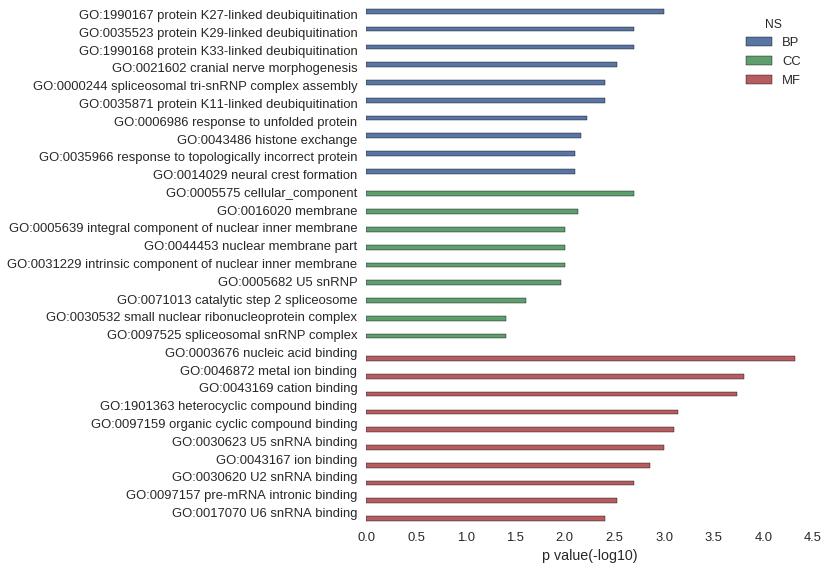
| GO | P value |
|---|---|
| GO:1990167 | 9.95e-04 |
| GO:0035523 | 1.99e-03 |
| GO:1990168 | 1.99e-03 |
| GO:0021602 | 2.98e-03 |
| GO:0000244 | 3.97e-03 |
| GO:0035871 | 3.97e-03 |
| GO:0006986 | 5.96e-03 |
| GO:0043486 | 6.94e-03 |
| GO:0035966 | 7.93e-03 |
| GO:0014029 | 7.93e-03 |
| GO:0005575 | 2.01e-03 |
| GO:0016020 | 7.34e-03 |
| GO:0005639 | 9.91e-03 |
| GO:0044453 | 9.91e-03 |
| GO:0031229 | 9.91e-03 |
| GO:0005682 | 1.09e-02 |
| GO:0071013 | 2.46e-02 |
| GO:0030532 | 3.91e-02 |
| GO:0097525 | 3.91e-02 |
| GO:0003676 | 4.73e-05 |
| GO:0046872 | 1.57e-04 |
| GO:0043169 | 1.85e-04 |
| GO:1901363 | 7.27e-04 |
| GO:0097159 | 7.92e-04 |
| GO:0030623 | 9.95e-04 |
| GO:0043167 | 1.37e-03 |
| GO:0030620 | 1.99e-03 |
| GO:0097157 | 2.98e-03 |
| GO:0017070 | 3.97e-03 |
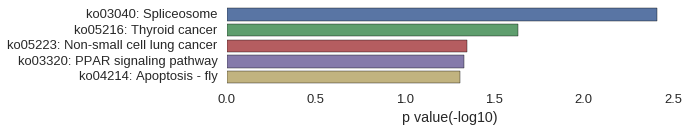
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02761
TTTGACAGCCCAGTGTACTGTCGTGATTGCATTCATTTCAATGGCTTTCTTCAAAATATTCTCTTTTTCG
GAAAAGTTGTGGGATTTAATCACGAAGGCGCGTGACGGTTCCTTGTCGTCCACTGGTTTGTTGCATAGAG
ATTGATTTGAGCGGGTAGACCTTTCCTCAAGATCTTGGCTTTGGTCTCAGCAGAGCCAGCTTCTTTTTTC
ACGATACTAAACTCTTTTTCCAGGGTAGTAAGAGAGTCGGAGTATCCACCCAACACAGGCTCCAGACCAG
TCACTCGCTCTTTCGCTGCTTCCACTCTGGCGTTAACCACTCTTCCAGTTCCTTTCTTGCTTTTTTCAAG
ATGGAGGTACGTGCACACGCTGTGGCTTTTCTCTTTCCGACAAGACATTGTTTTTGTGTTCTTTGTCATG
TTCGTCCTCGCCCCGTTTATCTGTGCTTAAGCGCACATATAGTCTAAAATTTTTGCTCGACATCGGCAGA
ACCAAACTTTACAAGTTAAACTCCGCAGACACGGAGGAGCTGCAAAATCTCCGTTTGCTCCAGAGGCCTA
CTCATCCTCCGACCCCCACCTCAACACCTCGCCCACAATGGAGGCAAGAGAAAGCAGAAGAGGGGCAAGC
GTGGGGGGATCTGAACTAGGCTAGCGGCTAACCCACAC
TTTGACAGCCCAGTGTACTGTCGTGATTGCATTCATTTCAATGGCTTTCTTCAAAATATTCTCTTTTTCG
GAAAAGTTGTGGGATTTAATCACGAAGGCGCGTGACGGTTCCTTGTCGTCCACTGGTTTGTTGCATAGAG
ATTGATTTGAGCGGGTAGACCTTTCCTCAAGATCTTGGCTTTGGTCTCAGCAGAGCCAGCTTCTTTTTTC
ACGATACTAAACTCTTTTTCCAGGGTAGTAAGAGAGTCGGAGTATCCACCCAACACAGGCTCCAGACCAG
TCACTCGCTCTTTCGCTGCTTCCACTCTGGCGTTAACCACTCTTCCAGTTCCTTTCTTGCTTTTTTCAAG
ATGGAGGTACGTGCACACGCTGTGGCTTTTCTCTTTCCGACAAGACATTGTTTTTGTGTTCTTTGTCATG
TTCGTCCTCGCCCCGTTTATCTGTGCTTAAGCGCACATATAGTCTAAAATTTTTGCTCGACATCGGCAGA
ACCAAACTTTACAAGTTAAACTCCGCAGACACGGAGGAGCTGCAAAATCTCCGTTTGCTCCAGAGGCCTA
CTCATCCTCCGACCCCCACCTCAACACCTCGCCCACAATGGAGGCAAGAGAAAGCAGAAGAGGGGCAAGC
GTGGGGGGATCTGAACTAGGCTAGCGGCTAACCCACAC