ZFLNCG02850
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516133 | skin | male and 5 month | 10.82 |
| SRR941749 | anterior pectoral fin | normal | 2.17 |
| SRR941753 | posterior pectoral fin | normal | 1.21 |
| SRR516131 | skin | male and 5 month | 0.70 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 0.64 |
| SRR516129 | skin | male and 5 month | 0.55 |
| SRR633516 | larvae | normal | 0.51 |
| SRR516127 | skin | male and 5 month | 0.50 |
| SRR516126 | skin | male and 5 month | 0.49 |
| SRR592703 | pineal gland | normal | 0.48 |
Express in tissues
Correlated coding gene
Download
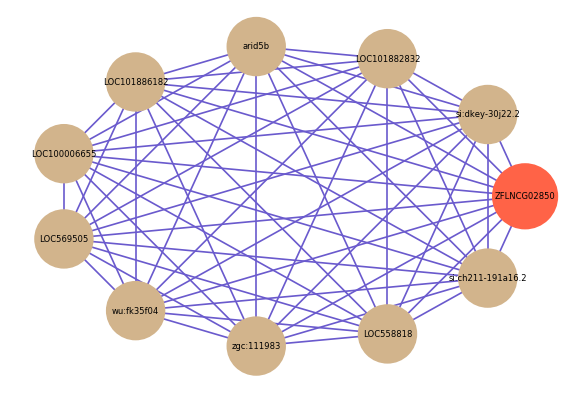
| gene | correlation coefficent |
|---|---|
| si:dkey-30j22.2 | 0.64 |
| LOC101882832 | 0.62 |
| arid5b | 0.61 |
| LOC101886182 | 0.60 |
| LOC100006655 | 0.60 |
| LOC569505 | 0.60 |
| wu:fk35f04 | 0.59 |
| zgc:111983 | 0.59 |
| LOC558818 | 0.59 |
| si:ch211-191a16.2 | 0.58 |
Gene Ontology
Download
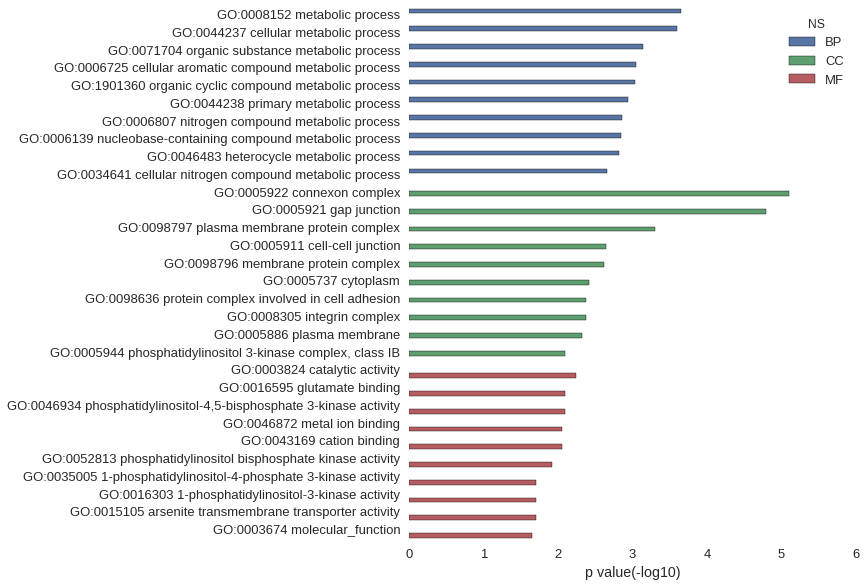
| GO | P value |
|---|---|
| GO:0008152 | 2.20e-04 |
| GO:0044237 | 2.52e-04 |
| GO:0071704 | 7.20e-04 |
| GO:0006725 | 9.09e-04 |
| GO:1901360 | 9.17e-04 |
| GO:0044238 | 1.14e-03 |
| GO:0006807 | 1.40e-03 |
| GO:0006139 | 1.43e-03 |
| GO:0046483 | 1.52e-03 |
| GO:0034641 | 2.22e-03 |
| GO:0005922 | 7.99e-06 |
| GO:0005921 | 1.61e-05 |
| GO:0098797 | 4.93e-04 |
| GO:0005911 | 2.28e-03 |
| GO:0098796 | 2.45e-03 |
| GO:0005737 | 3.81e-03 |
| GO:0098636 | 4.20e-03 |
| GO:0008305 | 4.20e-03 |
| GO:0005886 | 4.83e-03 |
| GO:0005944 | 8.09e-03 |
| GO:0003824 | 5.82e-03 |
| GO:0016595 | 8.09e-03 |
| GO:0046934 | 8.09e-03 |
| GO:0046872 | 8.75e-03 |
| GO:0043169 | 8.85e-03 |
| GO:0052813 | 1.21e-02 |
| GO:0035005 | 2.01e-02 |
| GO:0016303 | 2.01e-02 |
| GO:0015105 | 2.01e-02 |
| GO:0003674 | 2.22e-02 |
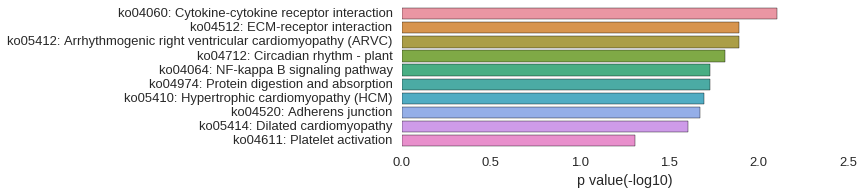
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02850
AGAATACTTGATCCTGAAGCAGTGAAACTGCACACTTATGAAGACACAAACATCACAGTATGTGTAGAGT
ATATTCAGTGAAATCAGCTGGAGAAAGTCTTTTATTGAGTCCAGCTGCTGTGAATGATATCAGGATCACA
CGACTTAACTGGGGCTAAAATTCACTCATTGTGACTTGTGTTACTGAGATTATAACAAGTACAATATAAA
GCACATTTGGAAAAGTGATCTGTTGTATCACAGCGACTGCTTTACTTTTGTACCGTCAACACTGCTATTA
CCAGCTGAGCAAAGCCTCCATTGCTATCTTCTATTCTTATCTATTTGTGTGTCTGTGTTTTTTTTTATCT
GACTGCTTATTTTCTCATGTCTCAAACAGGATGTGACGTGTTTTCCAGAGGTGAAAGAATTTTCAGATGT
GTTGTTCAGCTCAACAGTTCAGCGGAGAAATAGACAGATCTGCAGGCATTATTTGAGGTGAGAGTTTGCT
CTTTCTGTGCTTTATATCATCTATGAGTGTCGTCTGATTATCTGTCGATCATCTCTTTGTGTTTCTGAGA
CTTTACTGCTGAGCTGAAGACATTCTGAGGATTTTATTTACACCTTTTAAAGTCGAGTGTGTTTCTGGAA
GAGGATCACTGAAAGCAAAAACCTTCTCATCCTGATACTGAAATCTGACTTTATGAGTGTTTATTAAGCT
CAGCACAGTGGACATTTCTTTAGTTAAATCGTTTTAGTCGTTTGAAATGCAGTGACACACAATCAAATAA
TTCACCGAACAGATCAGCCACTTTTGACAAGATTCAGAGCACTGGAGCTTCTCAAACGTACAGGGAAACA
GGCCTGGCACAGTTCGGATAGCATAGTGTGTGTGTGCCCTTACAGAAGCTCTTAAATGCAGAAAACTCGC
ACTGCAATAACAGTATTAATGAAGGCATTTAGTTTATTTACAGTGGAATCACACAATCTAATGTGTTGAT
ACATGAGCTGAAAACCCTGAAGAGTTTCAGCAGTCAGACCGGTTTCATTTGTTTGCCTGAAGTGTTTCTG
CTTTAACTGATTCTGATGATAAACAACTGCTGTG
AGAATACTTGATCCTGAAGCAGTGAAACTGCACACTTATGAAGACACAAACATCACAGTATGTGTAGAGT
ATATTCAGTGAAATCAGCTGGAGAAAGTCTTTTATTGAGTCCAGCTGCTGTGAATGATATCAGGATCACA
CGACTTAACTGGGGCTAAAATTCACTCATTGTGACTTGTGTTACTGAGATTATAACAAGTACAATATAAA
GCACATTTGGAAAAGTGATCTGTTGTATCACAGCGACTGCTTTACTTTTGTACCGTCAACACTGCTATTA
CCAGCTGAGCAAAGCCTCCATTGCTATCTTCTATTCTTATCTATTTGTGTGTCTGTGTTTTTTTTTATCT
GACTGCTTATTTTCTCATGTCTCAAACAGGATGTGACGTGTTTTCCAGAGGTGAAAGAATTTTCAGATGT
GTTGTTCAGCTCAACAGTTCAGCGGAGAAATAGACAGATCTGCAGGCATTATTTGAGGTGAGAGTTTGCT
CTTTCTGTGCTTTATATCATCTATGAGTGTCGTCTGATTATCTGTCGATCATCTCTTTGTGTTTCTGAGA
CTTTACTGCTGAGCTGAAGACATTCTGAGGATTTTATTTACACCTTTTAAAGTCGAGTGTGTTTCTGGAA
GAGGATCACTGAAAGCAAAAACCTTCTCATCCTGATACTGAAATCTGACTTTATGAGTGTTTATTAAGCT
CAGCACAGTGGACATTTCTTTAGTTAAATCGTTTTAGTCGTTTGAAATGCAGTGACACACAATCAAATAA
TTCACCGAACAGATCAGCCACTTTTGACAAGATTCAGAGCACTGGAGCTTCTCAAACGTACAGGGAAACA
GGCCTGGCACAGTTCGGATAGCATAGTGTGTGTGTGCCCTTACAGAAGCTCTTAAATGCAGAAAACTCGC
ACTGCAATAACAGTATTAATGAAGGCATTTAGTTTATTTACAGTGGAATCACACAATCTAATGTGTTGAT
ACATGAGCTGAAAACCCTGAAGAGTTTCAGCAGTCAGACCGGTTTCATTTGTTTGCCTGAAGTGTTTCTG
CTTTAACTGATTCTGATGATAAACAACTGCTGTG