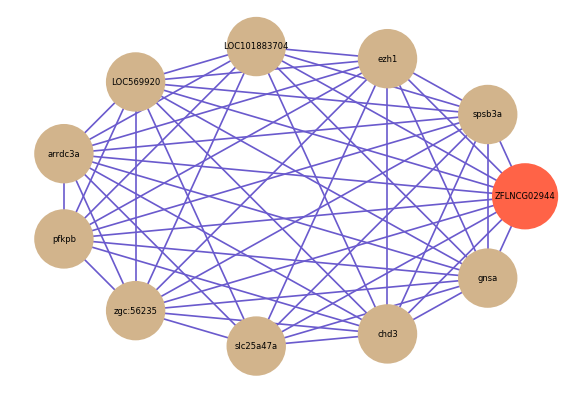
ZFLNCG02944
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 84.90 |
| SRR592703 | pineal gland | normal | 56.19 |
| SRR592701 | pineal gland | normal | 46.45 |
| SRR592702 | pineal gland | normal | 33.64 |
| ERR023146 | head kidney | normal | 32.85 |
| ERR023143 | swim bladder | normal | 28.76 |
| SRR592700 | pineal gland | normal | 24.31 |
| SRR592699 | pineal gland | normal | 24.10 |
| ERR023145 | heart | normal | 23.99 |
| SRR1648855 | brain | normal | 16.53 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
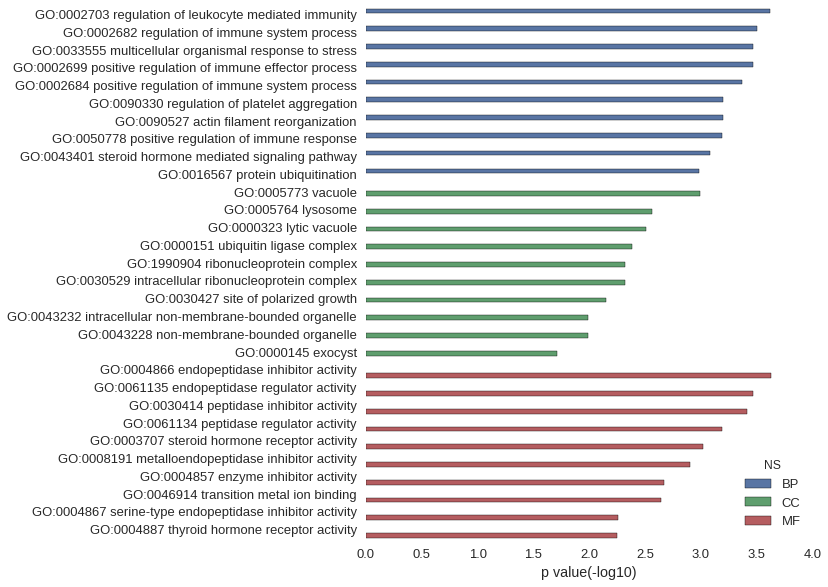
| GO | P value |
|---|---|
| GO:0002703 | 2.40e-04 |
| GO:0002682 | 3.11e-04 |
| GO:0033555 | 3.39e-04 |
| GO:0002699 | 3.39e-04 |
| GO:0002684 | 4.23e-04 |
| GO:0090330 | 6.28e-04 |
| GO:0090527 | 6.28e-04 |
| GO:0050778 | 6.43e-04 |
| GO:0043401 | 8.32e-04 |
| GO:0016567 | 1.03e-03 |
| GO:0005773 | 1.02e-03 |
| GO:0005764 | 2.72e-03 |
| GO:0000323 | 3.10e-03 |
| GO:0000151 | 4.13e-03 |
| GO:1990904 | 4.76e-03 |
| GO:0030529 | 4.76e-03 |
| GO:0030427 | 7.11e-03 |
| GO:0043232 | 1.02e-02 |
| GO:0043228 | 1.02e-02 |
| GO:0000145 | 1.96e-02 |
| GO:0004866 | 2.37e-04 |
| GO:0061135 | 3.38e-04 |
| GO:0030414 | 3.87e-04 |
| GO:0061134 | 6.41e-04 |
| GO:0003707 | 9.55e-04 |
| GO:0008191 | 1.24e-03 |
| GO:0004857 | 2.12e-03 |
| GO:0046914 | 2.28e-03 |
| GO:0004867 | 5.55e-03 |
| GO:0004887 | 5.58e-03 |
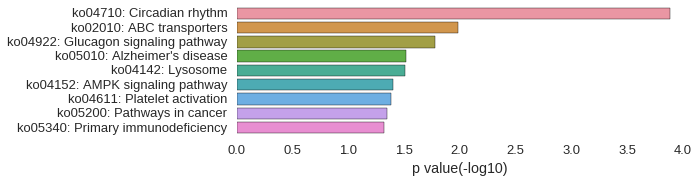
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02944
CTCGTCCACATACACTCCTACTCTCTTACAGGAGTCTGCGGACACACGCGTGTCGATGTGCTTGTAACCG
TACCAAAGACCGAATGTGTCGTCAGAGCAGATCAGGCACCAGGAGTTATCATTGAATCCAAACACACAGT
CAGCGTCCTTTCCTTTCCTGCCTATCCCTGCGTATGTGATGGCTATGTCAGCCTGTCCAGTCCATTCAGT
CTCCCAGTAGCAGCGTCCAGGCAGTGCTTCTCTAGACAGGACATGAGGACGCTTGTCAAATCTCTC
CTCGTCCACATACACTCCTACTCTCTTACAGGAGTCTGCGGACACACGCGTGTCGATGTGCTTGTAACCG
TACCAAAGACCGAATGTGTCGTCAGAGCAGATCAGGCACCAGGAGTTATCATTGAATCCAAACACACAGT
CAGCGTCCTTTCCTTTCCTGCCTATCCCTGCGTATGTGATGGCTATGTCAGCCTGTCCAGTCCATTCAGT
CTCCCAGTAGCAGCGTCCAGGCAGTGCTTCTCTAGACAGGACATGAGGACGCTTGTCAAATCTCTC