ZFLNCG02977
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR748488 | sperm | normal | 163.53 |
| ERR145648 | skeletal muscle | 27 degree_C to 27 degree_C | 6.59 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 3.52 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 3.04 |
| SRR748489 | egg | normal | 1.90 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 1.68 |
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 1.29 |
| SRR1021215 | sphere stage | Mzeomesa | 1.27 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 1.17 |
| SRR891504 | liver | normal | 0.93 |
Express in tissues
Correlated coding gene
Download
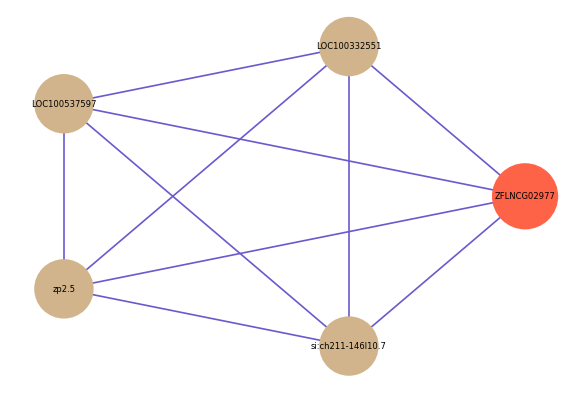
| gene | correlation coefficent |
|---|---|
| LOC100332551 | 0.52 |
| LOC100537597 | 0.52 |
| zp2.5 | 0.52 |
| si:ch211-146l10.7 | 0.51 |
Gene Ontology
Download

| GO | P value |
|---|---|
| GO:0005576 | 3.35e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02977
ACTGACTAGTAACAACTTTGGGCATTGATACAGCTACATATGGGGTTGAAGGTACTGAATCAACCTGGCT
TGTTGAACCTTTACCCTGGAGTAGTCTTTGCAGAGTGAAGGGAGCAGAAGAACCACTCGGAGTGCCATAG
ACAGAAGTAACCTGCTTTGAACTTGGCACTGGTGTCAACGAACTAGTAACAACTTGGGGCAATGATACAG
ATACGTATGGGGTTGAAGATTCATAAACAACCTGGCTTGTTGAATCTTGACCCTGGAGTAGTTCTTGCAA
AGTGAAAGGAGCAGGGGATCCTGTCTGGGTGGTATAGACAGAGGTAACCTGCTTTGTACTTGGTGCAGGT
GTCAATTGACTAGTTACTTGGGGCAATGATACAGCTACATATGGGGTTGAAGATCCTAAAACTAACGGAC
TTGTGGTATCTGGAGTCTCGATTCTTCCATCATATGTGGTACTTGCTTGCAAAGTGAAAGGGGCAGAGGG
TCCGGTCTGGGTGCCATAGACAGAGGTAACTTGCTTTGAACTTGGTCCAGCTGTCAGCTGGCTAGTAACA
TATTTGGGCAACGATATGGTTGCATATGGGGATGAAGACCCTGGAGCAAACGGACTTGTACCTTGGGTAG
TTACACCATAGGTGGCACTTTGAGAAACTGAACTAGACAGACCAGGGGAGCTCTGGTCATTAGTGGACTG
GCCAGAAACTGCAGTCTTGGAAGCAAAGCCGCCACTTCCCCCTTGTGCAAAGACAATTTGTCCAGAACCA
CTTGTAGTGGTGGATCCAGAGGAGCCCTGAAGTGACACTACACCACTCATCCCAGATTGATCCACAGGCT
GAGTAGCTGAAGACGGGGTATTTGGAGGCAGAGCTTGGGGTGTGGAGTCAGAACTGCTGGAAACACCACC
TTTAACAACCTGAAGCAGATTAAACTTGGGGTTGTTTTTTGGCAAACGAGAGACCACTTCTTCAAGCAAA
ACCCCAGTCACTGGGGCTTCTGGCACAGAAGACTGGACCACAAGAGGCAAGAAAGCGTTGTCAGATGG
ACTGACTAGTAACAACTTTGGGCATTGATACAGCTACATATGGGGTTGAAGGTACTGAATCAACCTGGCT
TGTTGAACCTTTACCCTGGAGTAGTCTTTGCAGAGTGAAGGGAGCAGAAGAACCACTCGGAGTGCCATAG
ACAGAAGTAACCTGCTTTGAACTTGGCACTGGTGTCAACGAACTAGTAACAACTTGGGGCAATGATACAG
ATACGTATGGGGTTGAAGATTCATAAACAACCTGGCTTGTTGAATCTTGACCCTGGAGTAGTTCTTGCAA
AGTGAAAGGAGCAGGGGATCCTGTCTGGGTGGTATAGACAGAGGTAACCTGCTTTGTACTTGGTGCAGGT
GTCAATTGACTAGTTACTTGGGGCAATGATACAGCTACATATGGGGTTGAAGATCCTAAAACTAACGGAC
TTGTGGTATCTGGAGTCTCGATTCTTCCATCATATGTGGTACTTGCTTGCAAAGTGAAAGGGGCAGAGGG
TCCGGTCTGGGTGCCATAGACAGAGGTAACTTGCTTTGAACTTGGTCCAGCTGTCAGCTGGCTAGTAACA
TATTTGGGCAACGATATGGTTGCATATGGGGATGAAGACCCTGGAGCAAACGGACTTGTACCTTGGGTAG
TTACACCATAGGTGGCACTTTGAGAAACTGAACTAGACAGACCAGGGGAGCTCTGGTCATTAGTGGACTG
GCCAGAAACTGCAGTCTTGGAAGCAAAGCCGCCACTTCCCCCTTGTGCAAAGACAATTTGTCCAGAACCA
CTTGTAGTGGTGGATCCAGAGGAGCCCTGAAGTGACACTACACCACTCATCCCAGATTGATCCACAGGCT
GAGTAGCTGAAGACGGGGTATTTGGAGGCAGAGCTTGGGGTGTGGAGTCAGAACTGCTGGAAACACCACC
TTTAACAACCTGAAGCAGATTAAACTTGGGGTTGTTTTTTGGCAAACGAGAGACCACTTCTTCAAGCAAA
ACCCCAGTCACTGGGGCTTCTGGCACAGAAGACTGGACCACAAGAGGCAAGAAAGCGTTGTCAGATGG
