ZFLNCG03001
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 25.58 |
| ERR023146 | head kidney | normal | 17.47 |
| SRR516122 | skin | male and 3.5 year | 9.28 |
| ERR023145 | heart | normal | 8.41 |
| SRR891511 | brain | normal | 7.58 |
| SRR038627 | embryo | control morpholino | 6.91 |
| SRR516123 | skin | male and 3.5 year | 6.06 |
| SRR726542 | 5 dpf | infection with control | 6.01 |
| ERR023143 | swim bladder | normal | 5.79 |
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 3.27 |
Express in tissues
Correlated coding gene
Download
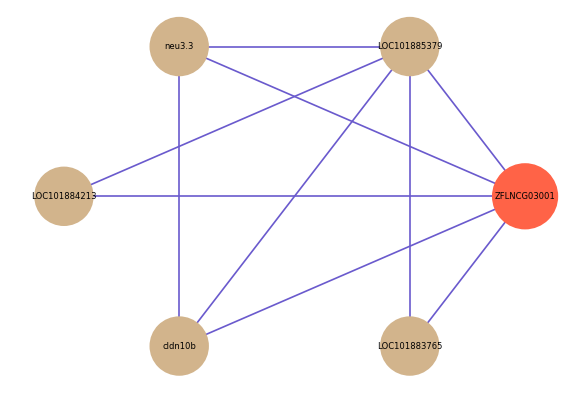
| gene | correlation coefficent |
|---|---|
| LOC101885379 | 0.57 |
| neu3.3 | 0.53 |
| LOC101884213 | 0.52 |
| cldn10b | 0.52 |
| LOC101883765 | 0.50 |
Gene Ontology
Download
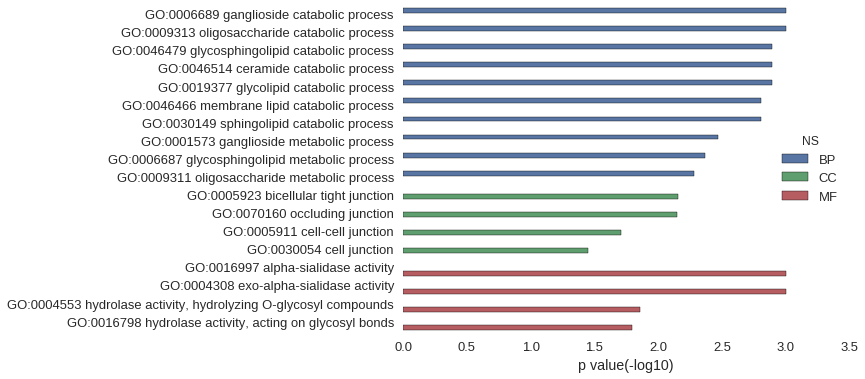
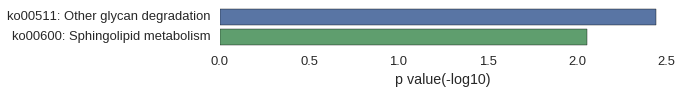
| GO | P value |
|---|---|
| GO:0006689 | 9.95e-04 |
| GO:0009313 | 9.95e-04 |
| GO:0046479 | 1.28e-03 |
| GO:0046514 | 1.28e-03 |
| GO:0019377 | 1.28e-03 |
| GO:0046466 | 1.56e-03 |
| GO:0030149 | 1.56e-03 |
| GO:0001573 | 3.41e-03 |
| GO:0006687 | 4.26e-03 |
| GO:0009311 | 5.25e-03 |
| GO:0005923 | 6.95e-03 |
| GO:0070160 | 7.09e-03 |
| GO:0005911 | 1.94e-02 |
| GO:0030054 | 3.56e-02 |
| GO:0016997 | 9.95e-04 |
| GO:0004308 | 9.95e-04 |
| GO:0004553 | 1.39e-02 |
| GO:0016798 | 1.59e-02 |
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT04584 | NONMMUT109455; | Direct blastn |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03001
TAATGTTGGGTGTTATCGTACAGACACTTCAACACTCGCTGTTTCTAGAAGATCTAGCAGGGCATCTGGG
CAGCTTCAGTGTACATTAGTCTGCCGCACACTGATTATATGTATCAACACTAGTTAGGGTGGATAGTGTG
TGAATTGAGGCGCAGCCGGAGTGTCTGAAGTGTGCATCCAAAGTACACAAATAGCAGCAAGTGTGTCCTC
ACTTCTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGGTGTTCTGCTGCTTCTGAAATGTGTGT
GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTTTGAG
TAATGTTGGGTGTTATCGTACAGACACTTCAACACTCGCTGTTTCTAGAAGATCTAGCAGGGCATCTGGG
CAGCTTCAGTGTACATTAGTCTGCCGCACACTGATTATATGTATCAACACTAGTTAGGGTGGATAGTGTG
TGAATTGAGGCGCAGCCGGAGTGTCTGAAGTGTGCATCCAAAGTACACAAATAGCAGCAAGTGTGTCCTC
ACTTCTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGGTGTTCTGCTGCTTCTGAAATGTGTGT
GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTTTGAG