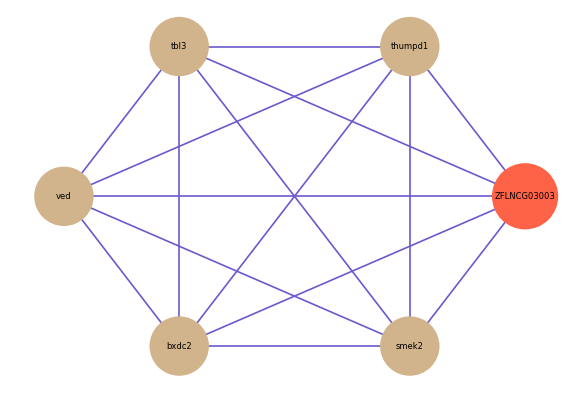
ZFLNCG03003
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 2.73 |
| SRR891495 | heart | normal | 2.63 |
| SRR038625 | embryo | traf6 morpholino | 2.13 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 1.79 |
| SRR891512 | blood | normal | 1.79 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 1.56 |
| SRR038627 | embryo | control morpholino | 1.44 |
| SRR658537 | bud | Gata6 morphant | 1.37 |
| SRR1299125 | caudal fin | Half day time after treatment | 1.36 |
| SRR658543 | 6 somite | Gata5 morphan | 1.28 |
Express in tissues
Correlated coding gene
Download
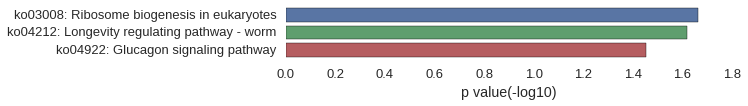
Gene Ontology
Download
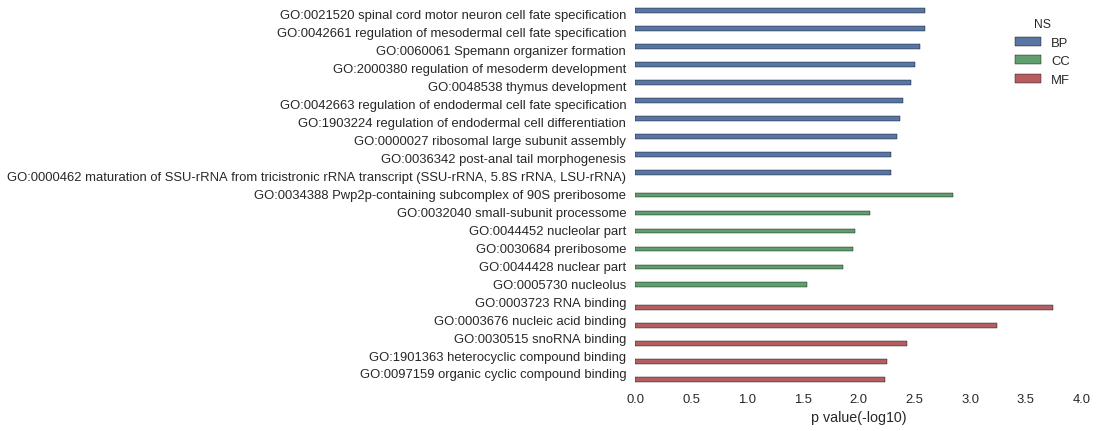
| GO | P value |
|---|---|
| GO:0021520 | 2.56e-03 |
| GO:0042661 | 2.56e-03 |
| GO:0060061 | 2.84e-03 |
| GO:2000380 | 3.12e-03 |
| GO:0048538 | 3.41e-03 |
| GO:0042663 | 3.97e-03 |
| GO:1903224 | 4.26e-03 |
| GO:0000027 | 4.54e-03 |
| GO:0036342 | 5.11e-03 |
| GO:0000462 | 5.11e-03 |
| GO:0034388 | 1.42e-03 |
| GO:0032040 | 7.94e-03 |
| GO:0044452 | 1.08e-02 |
| GO:0030684 | 1.13e-02 |
| GO:0044428 | 1.39e-02 |
| GO:0005730 | 2.87e-02 |
| GO:0003723 | 1.79e-04 |
| GO:0003676 | 5.69e-04 |
| GO:0030515 | 3.69e-03 |
| GO:1901363 | 5.52e-03 |
| GO:0097159 | 5.74e-03 |
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT04587 | NONHSAT154332; | Direct blastn |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03003
TGGTTTTGTGAATGATGAATGTCCTGAAAGAGATGTTGGTCATTACCCCACACACAGTTTATACCAAACT
AAATAAGAGACGTCACCATTAAACTGATTCACAGATGCTCTCCAGACAGGGGTAATCTGAACAGCCTTGA
GAATGACCTGAACAGCAGAGCTCTGAAGGACTGTGGTGAAGCGCTGCCACTTGTCTTGGACTGAATGGAC
GTTTGAGACTGCAGCTCTGACACCAGTGAGCAGCATTTACACACACTCACTACATCATCAACACATTACT
AATCAATACAGCAGCTTTACTAGTTCACAAGCCCACTCTGTATTAGTTTAAATGCATATGTAACATTATT
TAATATAAAATAGTTTATATTTATACTTATTCAAGAAAATAACATCATGTAAAACTAAAAAACGCAAGGT
GTTTTTCAGTGTGTGTGCGTCTGCATGTGTACAAGTGTTCATGAGTTATTGTGTGTGTGTGTGTGTGTGT
GTGTGTGTGTGTGTGTGTGTGTGTGCGGTGCTGCTGCCCTCTGCTGGTCATCTGTGCTCCTGCAGTCGCC
CTCCTCCTCTGTCTATCATCAGTATAGATCAGGGGGTTTCCTAAACCTTTCCACCTGCGAGCCCTAAAAC
AACACTGCCAGTAACTCGCGACCCCCACATTCCTCTGAGGTGGTTATAAATATGTTGTGCACACAATCGT
CTGCTGATATAAACACCAGCAGACTGACAACACAAA
TGGTTTTGTGAATGATGAATGTCCTGAAAGAGATGTTGGTCATTACCCCACACACAGTTTATACCAAACT
AAATAAGAGACGTCACCATTAAACTGATTCACAGATGCTCTCCAGACAGGGGTAATCTGAACAGCCTTGA
GAATGACCTGAACAGCAGAGCTCTGAAGGACTGTGGTGAAGCGCTGCCACTTGTCTTGGACTGAATGGAC
GTTTGAGACTGCAGCTCTGACACCAGTGAGCAGCATTTACACACACTCACTACATCATCAACACATTACT
AATCAATACAGCAGCTTTACTAGTTCACAAGCCCACTCTGTATTAGTTTAAATGCATATGTAACATTATT
TAATATAAAATAGTTTATATTTATACTTATTCAAGAAAATAACATCATGTAAAACTAAAAAACGCAAGGT
GTTTTTCAGTGTGTGTGCGTCTGCATGTGTACAAGTGTTCATGAGTTATTGTGTGTGTGTGTGTGTGTGT
GTGTGTGTGTGTGTGTGTGTGTGTGCGGTGCTGCTGCCCTCTGCTGGTCATCTGTGCTCCTGCAGTCGCC
CTCCTCCTCTGTCTATCATCAGTATAGATCAGGGGGTTTCCTAAACCTTTCCACCTGCGAGCCCTAAAAC
AACACTGCCAGTAACTCGCGACCCCCACATTCCTCTGAGGTGGTTATAAATATGTTGTGCACACAATCGT
CTGCTGATATAAACACCAGCAGACTGACAACACAAA