ZFLNCG03013
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR527834 | head | normal | 9.81 |
| SRR527835 | tail | normal | 7.93 |
| SRR658537 | bud | Gata6 morphant | 5.86 |
| SRR658547 | 6 somite | Gata5/6 morphant | 5.45 |
| SRR527833 | 5 dpf | normal | 5.10 |
| SRR658543 | 6 somite | Gata5 morphan | 5.01 |
| SRR658533 | bud | normal | 4.62 |
| SRR800045 | muscle | normal | 4.00 |
| SRR658539 | bud | Gata5/6 morphant | 3.79 |
| SRR658535 | bud | Gata5 morphant | 3.57 |
Express in tissues
Correlated coding gene
Download
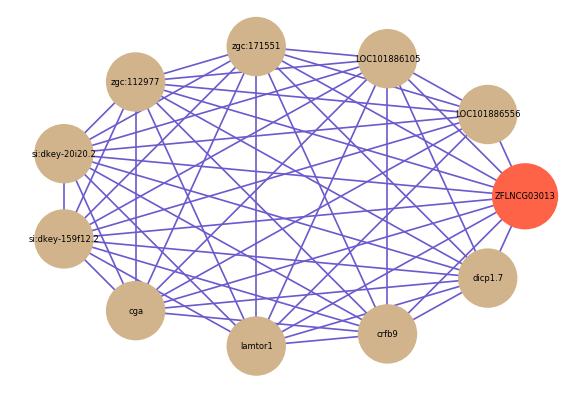
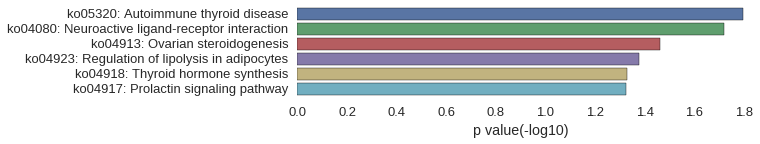
| gene | correlation coefficent |
|---|---|
| LOC101886556 | 0.58 |
| LOC101886105 | 0.57 |
| zgc:171551 | 0.56 |
| crfb9 | 0.56 |
| dicp1.7 | 0.55 |
| zgc:112977 | 0.54 |
| si:dkey-20i20.2 | 0.54 |
| si:dkey-159f12.2 | 0.54 |
| cga | 0.53 |
| lamtor1 | 0.53 |
Gene Ontology
Download
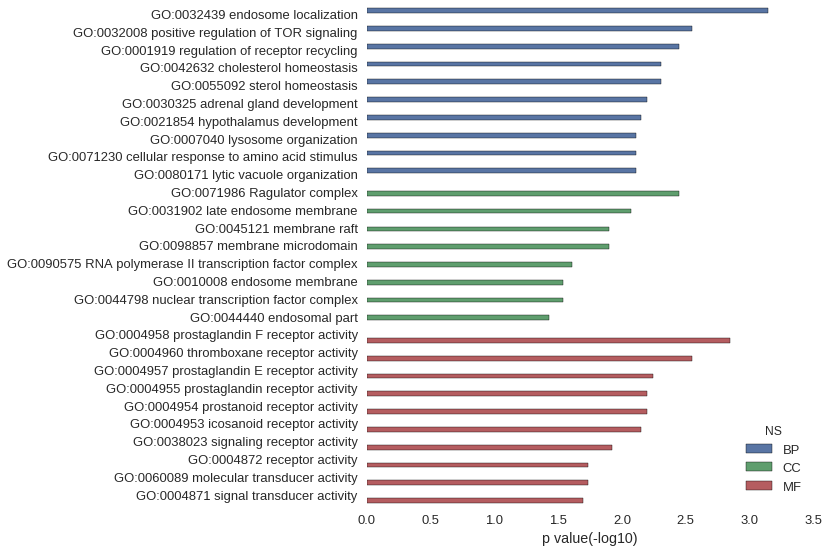
| GO | P value |
|---|---|
| GO:0032439 | 7.11e-04 |
| GO:0032008 | 2.84e-03 |
| GO:0001919 | 3.55e-03 |
| GO:0042632 | 4.96e-03 |
| GO:0055092 | 4.96e-03 |
| GO:0030325 | 6.38e-03 |
| GO:0021854 | 7.09e-03 |
| GO:0007040 | 7.79e-03 |
| GO:0071230 | 7.79e-03 |
| GO:0080171 | 7.79e-03 |
| GO:0071986 | 3.55e-03 |
| GO:0031902 | 8.50e-03 |
| GO:0045121 | 1.27e-02 |
| GO:0098857 | 1.27e-02 |
| GO:0090575 | 2.46e-02 |
| GO:0010008 | 2.88e-02 |
| GO:0044798 | 2.88e-02 |
| GO:0044440 | 3.70e-02 |
| GO:0004958 | 1.42e-03 |
| GO:0004960 | 2.84e-03 |
| GO:0004957 | 5.67e-03 |
| GO:0004955 | 6.38e-03 |
| GO:0004954 | 6.38e-03 |
| GO:0004953 | 7.09e-03 |
| GO:0038023 | 1.20e-02 |
| GO:0004872 | 1.85e-02 |
| GO:0060089 | 1.85e-02 |
| GO:0004871 | 2.01e-02 |
KEGG Pathway
Download
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT04607 | NONMMUT113394; | Direct blastn |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03013
TTTCTGTGTGTGTGTGTGTGTGTTTCTGCAGTTGGTGGACAAGCTGAACACTAACACTGAGATGGAGGAA
GTGGTGAATGACATCATCATGGTGAGCAAATCAGCACTGGACACTTCCTGTTCAGCGTCCTGCCATTCTG
CTGATGTTCTGCGCGTGTGTGTGTGTGTGTGTGTGTGTGTGTTTTCTCCAGCTGAAGGCGCAGAACGACA
GAGAAGCTCAGAGTATAGACGTGATCTTCACACAGAGACGACAG
TTTCTGTGTGTGTGTGTGTGTGTTTCTGCAGTTGGTGGACAAGCTGAACACTAACACTGAGATGGAGGAA
GTGGTGAATGACATCATCATGGTGAGCAAATCAGCACTGGACACTTCCTGTTCAGCGTCCTGCCATTCTG
CTGATGTTCTGCGCGTGTGTGTGTGTGTGTGTGTGTGTGTGTTTTCTCCAGCTGAAGGCGCAGAACGACA
GAGAAGCTCAGAGTATAGACGTGATCTTCACACAGAGACGACAG