ZFLNCG03067
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1621206 | embryo | normal | 563.74 |
| SRR1049814 | 256 cell | normal | 484.17 |
| SRR372787 | 2-4 cell | normal | 348.17 |
| SRR372793 | shield | normal | 239.96 |
| SRR372791 | dome | normal | 178.78 |
| SRR372789 | 1K cell | normal | 136.16 |
| ERR023145 | heart | normal | 95.15 |
| SRR372802 | 5 dpf | normal | 73.11 |
| SRR748489 | egg | normal | 40.09 |
| SRR592698 | pineal gland | normal | 39.00 |
Express in tissues
Gene Ontology
Download
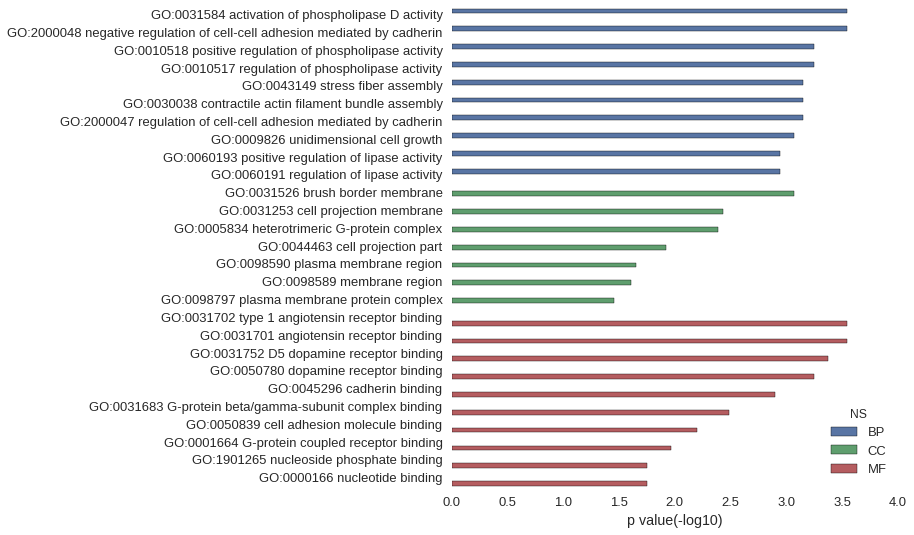
| GO | P value |
|---|---|
| GO:0031584 | 2.84e-04 |
| GO:2000048 | 2.84e-04 |
| GO:0010518 | 5.68e-04 |
| GO:0010517 | 5.68e-04 |
| GO:0043149 | 7.11e-04 |
| GO:0030038 | 7.11e-04 |
| GO:2000047 | 7.11e-04 |
| GO:0009826 | 8.53e-04 |
| GO:0060193 | 1.14e-03 |
| GO:0060191 | 1.14e-03 |
| GO:0031526 | 8.53e-04 |
| GO:0031253 | 3.69e-03 |
| GO:0005834 | 4.12e-03 |
| GO:0044463 | 1.20e-02 |
| GO:0098590 | 2.23e-02 |
| GO:0098589 | 2.47e-02 |
| GO:0098797 | 3.51e-02 |
| GO:0031702 | 2.84e-04 |
| GO:0031701 | 2.84e-04 |
| GO:0031752 | 4.26e-04 |
| GO:0050780 | 5.68e-04 |
| GO:0045296 | 1.28e-03 |
| GO:0031683 | 3.27e-03 |
| GO:0050839 | 6.39e-03 |
| GO:0001664 | 1.08e-02 |
| GO:1901265 | 1.76e-02 |
| GO:0000166 | 1.76e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03067
GTCAACAGGCCAGTACGCGGGCGCGTGTCCGACTCCTCACGTGCTCGCGCTGCGCGTGCACTTGTTGAAA
ACTTCTCCGCGTGCAATTGCGGAGCACAGACGAGCCACGAAACAGAGTCGAGCGGTAAATCGCGGGCTCC
TGCTCCCGTTATCGACCCAGAACGACCGCGAAACATGGACGATTTAGGTAAGCGTGATCTCCTTTAACCT
TTGACATCGGAAAATTGTGAAATGGGAAGTTTGACGCGACGCTAGAGCGCTAAAATGTGGCTGAAGACGC
GGATGGAGCGCGCTCCTGTTGCCAACGTCATCGCGCGCCGATGCTTGACTGCAGCAACACGCTGAAAAAC
ATACTCTCTGCTGCGGTTTAAACCTGGGTTTATTCGCGTATTTTACTCTGTTTTCTGTAGCTTCTCAATG
GGTTGTCGGAGTCAGCCTGATTCATTGTTCACATTGAGGTGAAGTTGGTGTTATTCACGTATTCGTTCGT
TTTTTTAACAGTGAAAACTTGTGAAGCTACTTTGGGCACTCAATCTGAAATCGCATTTAAGTAACTTATG
ACTTCAAAACAACATTAGCACCATTACATTAACTGTGTGTGAACACTATTGTGCTTTGATACTCATTGGC
TGCTAATATGATTGAGATTTTGCTAATAATACTTTTCTGGATTCATATTTTTAAGGAGAATGAGCAAATA
TAAAACCAACTTTACCTCAATGCTCTTCACAGCTGCTCTTGTAACACGAAGATACGTGATTAATATCAGA
TCCTCACCAGCGCAAA
GTCAACAGGCCAGTACGCGGGCGCGTGTCCGACTCCTCACGTGCTCGCGCTGCGCGTGCACTTGTTGAAA
ACTTCTCCGCGTGCAATTGCGGAGCACAGACGAGCCACGAAACAGAGTCGAGCGGTAAATCGCGGGCTCC
TGCTCCCGTTATCGACCCAGAACGACCGCGAAACATGGACGATTTAGGTAAGCGTGATCTCCTTTAACCT
TTGACATCGGAAAATTGTGAAATGGGAAGTTTGACGCGACGCTAGAGCGCTAAAATGTGGCTGAAGACGC
GGATGGAGCGCGCTCCTGTTGCCAACGTCATCGCGCGCCGATGCTTGACTGCAGCAACACGCTGAAAAAC
ATACTCTCTGCTGCGGTTTAAACCTGGGTTTATTCGCGTATTTTACTCTGTTTTCTGTAGCTTCTCAATG
GGTTGTCGGAGTCAGCCTGATTCATTGTTCACATTGAGGTGAAGTTGGTGTTATTCACGTATTCGTTCGT
TTTTTTAACAGTGAAAACTTGTGAAGCTACTTTGGGCACTCAATCTGAAATCGCATTTAAGTAACTTATG
ACTTCAAAACAACATTAGCACCATTACATTAACTGTGTGTGAACACTATTGTGCTTTGATACTCATTGGC
TGCTAATATGATTGAGATTTTGCTAATAATACTTTTCTGGATTCATATTTTTAAGGAGAATGAGCAAATA
TAAAACCAACTTTACCTCAATGCTCTTCACAGCTGCTCTTGTAACACGAAGATACGTGATTAATATCAGA
TCCTCACCAGCGCAAA