ZFLNCG03262
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 6.48 |
| SRR592700 | pineal gland | normal | 5.16 |
| ERR023143 | swim bladder | normal | 4.41 |
| ERR023144 | brain | normal | 4.17 |
| SRR516129 | skin | male and 5 month | 2.74 |
| SRR1534351 | larvae | 500 ug/l BDE47 treatment | 2.35 |
| SRR630463 | 5 dpf | normal | 1.90 |
| SRR1048061 | pineal gland | dark | 1.80 |
| SRR592703 | pineal gland | normal | 1.69 |
| ERR594416 | retina | transgenic flk1 | 1.68 |
Express in tissues
Correlated coding gene
Download
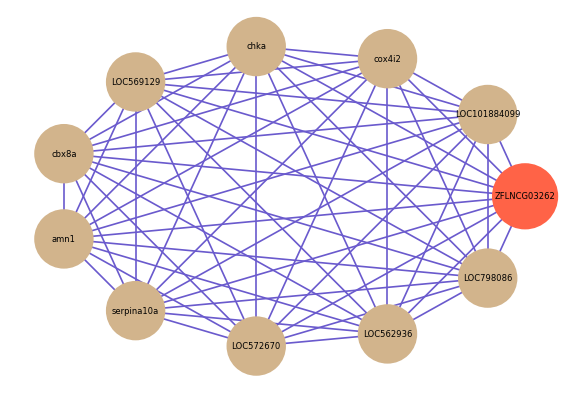
| gene | correlation coefficent |
|---|---|
| LOC101884099 | 0.53 |
| cox4i2 | 0.52 |
| chka | 0.52 |
| LOC569129 | 0.52 |
| cbx8a | 0.51 |
| amn1 | 0.51 |
| serpina10a | 0.51 |
| LOC572670 | 0.51 |
| LOC562936 | 0.50 |
| LOC798086 | 0.50 |
Gene Ontology
Download
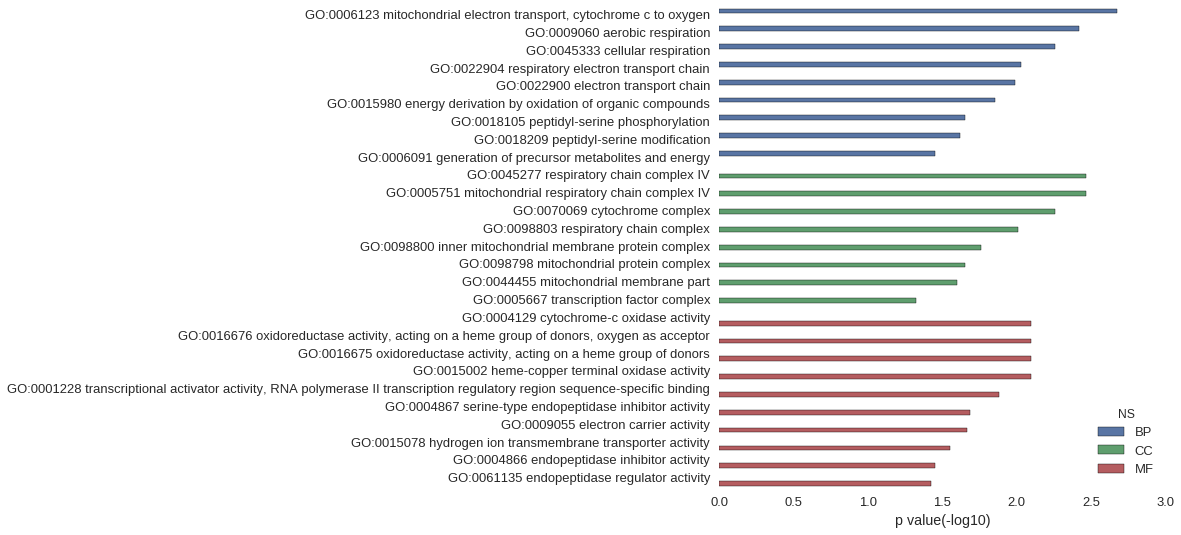
| GO | P value |
|---|---|
| GO:0006123 | 2.13e-03 |
| GO:0009060 | 3.83e-03 |
| GO:0045333 | 5.53e-03 |
| GO:0022904 | 9.35e-03 |
| GO:0022900 | 1.02e-02 |
| GO:0015980 | 1.40e-02 |
| GO:0018105 | 2.24e-02 |
| GO:0018209 | 2.41e-02 |
| GO:0006091 | 3.57e-02 |
| GO:0045277 | 3.41e-03 |
| GO:0005751 | 3.41e-03 |
| GO:0070069 | 5.53e-03 |
| GO:0098803 | 9.77e-03 |
| GO:0098800 | 1.74e-02 |
| GO:0098798 | 2.24e-02 |
| GO:0044455 | 2.53e-02 |
| GO:0005667 | 4.76e-02 |
| GO:0004129 | 8.08e-03 |
| GO:0016676 | 8.08e-03 |
| GO:0016675 | 8.08e-03 |
| GO:0015002 | 8.08e-03 |
| GO:0001228 | 1.31e-02 |
| GO:0004867 | 2.07e-02 |
| GO:0009055 | 2.16e-02 |
| GO:0015078 | 2.82e-02 |
| GO:0004866 | 3.57e-02 |
| GO:0061135 | 3.78e-02 |
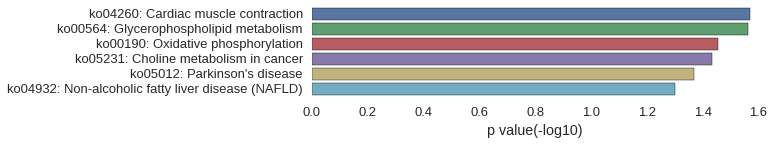
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03262
CCATTCTGGTCATAATGTGTTTTCATTGCCTTTAAAATATTTTAAAAACACAAATCATACATATTAAGTC
TGTATAAATGTATAACAGTGTGTTATTTGACTTATAATATACGCCATGGAGAGATTTTACACATGCAGTT
GCTTTTTCACAGACAATATTTTTCTTGAAGAATACAAACTTCATGTTCGTTTCGTCTCAGAGAACCAGTT
TAGGAAGGACTATCAAAATGTAAGTAATTTACAGCAAGACGGGCTCATGTCTTCATGTTAAACAGTTTGC
CAATTCCGTCTGAACTTCGCTGACATTTGCTTAAGATCCTCAGATCCCTGGGTTGTGAGTCTGAATCCCA
ATTCAGAGACGTGATCGGAAAGGTGAGTAATAAGTGTGCTCTCGAGAACAGCGATAACGATGAAGATCAA
GGCTGTCAGAGAGCGGTTATTCACTCACTCGTGTATTTATAGATTCAGGCAGAAATCGAAAGACGCCAAA
ATCATAAATTGAAATCTACAGAGAGAGCAGCTGTCATCAAGGAGA
CCATTCTGGTCATAATGTGTTTTCATTGCCTTTAAAATATTTTAAAAACACAAATCATACATATTAAGTC
TGTATAAATGTATAACAGTGTGTTATTTGACTTATAATATACGCCATGGAGAGATTTTACACATGCAGTT
GCTTTTTCACAGACAATATTTTTCTTGAAGAATACAAACTTCATGTTCGTTTCGTCTCAGAGAACCAGTT
TAGGAAGGACTATCAAAATGTAAGTAATTTACAGCAAGACGGGCTCATGTCTTCATGTTAAACAGTTTGC
CAATTCCGTCTGAACTTCGCTGACATTTGCTTAAGATCCTCAGATCCCTGGGTTGTGAGTCTGAATCCCA
ATTCAGAGACGTGATCGGAAAGGTGAGTAATAAGTGTGCTCTCGAGAACAGCGATAACGATGAAGATCAA
GGCTGTCAGAGAGCGGTTATTCACTCACTCGTGTATTTATAGATTCAGGCAGAAATCGAAAGACGCCAAA
ATCATAAATTGAAATCTACAGAGAGAGCAGCTGTCATCAAGGAGA