ZFLNCG03447
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592700 | pineal gland | normal | 52.07 |
| SRR592701 | pineal gland | normal | 41.16 |
| SRR1299127 | caudal fin | Two days time after treatment | 26.16 |
| SRR1299126 | caudal fin | One day time after treatment | 21.74 |
| SRR941753 | posterior pectoral fin | normal | 20.22 |
| SRR1299124 | caudal fin | Zero day time after treatment | 19.42 |
| SRR941749 | anterior pectoral fin | normal | 18.23 |
| SRR1299128 | caudal fin | Three days time after treatment | 17.63 |
| SRR1299125 | caudal fin | Half day time after treatment | 17.44 |
| SRR1562529 | intestine and pancreas | normal | 16.99 |
Express in tissues
Correlated coding gene
Download
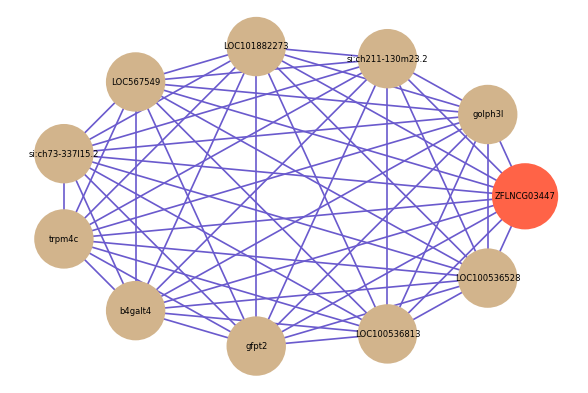
| gene | correlation coefficent |
|---|---|
| golph3l | 0.64 |
| si:ch211-130m23.2 | 0.61 |
| LOC101882273 | 0.61 |
| LOC567549 | 0.61 |
| si:ch73-337l15.2 | 0.61 |
| trpm4c | 0.60 |
| b4galt4 | 0.60 |
| gfpt2 | 0.60 |
| LOC100536813 | 0.60 |
| LOC100536528 | 0.60 |
Gene Ontology
Download
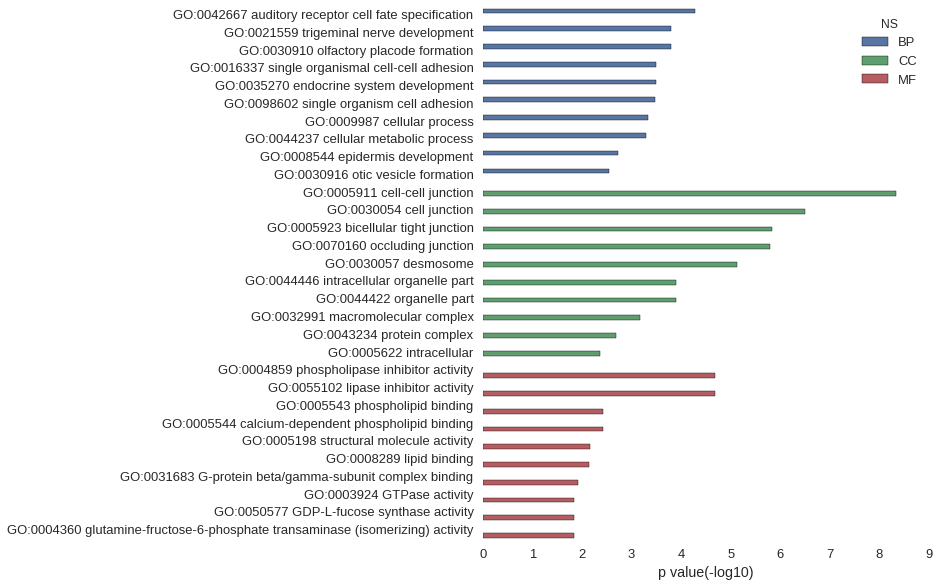
| GO | P value |
|---|---|
| GO:0042667 | 5.31e-05 |
| GO:0021559 | 1.58e-04 |
| GO:0030910 | 1.58e-04 |
| GO:0016337 | 3.14e-04 |
| GO:0035270 | 3.15e-04 |
| GO:0098602 | 3.33e-04 |
| GO:0009987 | 4.62e-04 |
| GO:0044237 | 5.06e-04 |
| GO:0008544 | 1.85e-03 |
| GO:0030916 | 2.80e-03 |
| GO:0005911 | 4.62e-09 |
| GO:0030054 | 3.16e-07 |
| GO:0005923 | 1.44e-06 |
| GO:0070160 | 1.63e-06 |
| GO:0030057 | 7.50e-06 |
| GO:0044446 | 1.29e-04 |
| GO:0044422 | 1.29e-04 |
| GO:0032991 | 6.73e-04 |
| GO:0043234 | 2.03e-03 |
| GO:0005622 | 4.32e-03 |
| GO:0004859 | 2.08e-05 |
| GO:0055102 | 2.08e-05 |
| GO:0005543 | 3.75e-03 |
| GO:0005544 | 3.80e-03 |
| GO:0005198 | 6.72e-03 |
| GO:0008289 | 7.28e-03 |
| GO:0031683 | 1.21e-02 |
| GO:0003924 | 1.41e-02 |
| GO:0050577 | 1.46e-02 |
| GO:0004360 | 1.46e-02 |
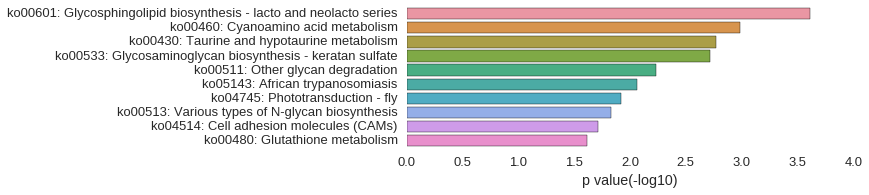
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03447
CTGTTTTCACTTTGCACTTTACGTGTCTGCCCATTTCACTGGCCTGATTTGACAAGGGGGCTGAGTCAAG
GGAACCTGTGTGTTCACGGCAGGCCCAGACCTGTCACCATTTAAGCAACGTCATAAACTGGAGACACTGT
TACACTGAGGTGGTCTTACAGCTCAGGCCTGCTGTAAACTTCCAAGTTCCCAACATAAGTGCAAAGTGTG
AACAGCATCGCAA
CTGTTTTCACTTTGCACTTTACGTGTCTGCCCATTTCACTGGCCTGATTTGACAAGGGGGCTGAGTCAAG
GGAACCTGTGTGTTCACGGCAGGCCCAGACCTGTCACCATTTAAGCAACGTCATAAACTGGAGACACTGT
TACACTGAGGTGGTCTTACAGCTCAGGCCTGCTGTAAACTTCCAAGTTCCCAACATAAGTGCAAAGTGTG
AACAGCATCGCAA