ZFLNCG03692
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 156.57 |
| ERR023145 | heart | normal | 154.34 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 152.52 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 148.26 |
| SRR658533 | bud | normal | 96.34 |
| SRR658535 | bud | Gata5 morphant | 90.08 |
| SRR658539 | bud | Gata5/6 morphant | 89.81 |
| SRR891495 | heart | normal | 84.64 |
| SRR1299125 | caudal fin | Half day time after treatment | 80.80 |
| SRR658537 | bud | Gata6 morphant | 76.94 |
Express in tissues
Correlated coding gene
Download
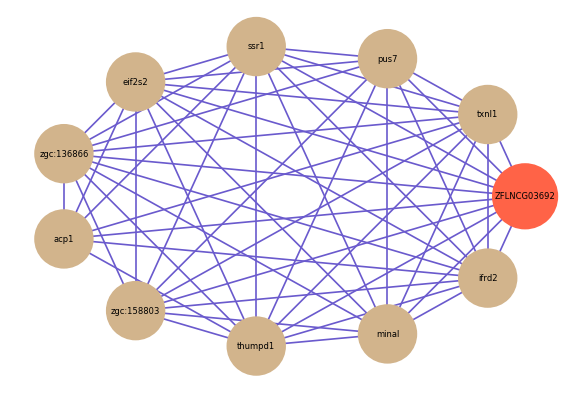
| gene | correlation coefficent |
|---|---|
| txnl1 | 0.65 |
| pus7 | 0.65 |
| ssr1 | 0.64 |
| eif2s2 | 0.64 |
| zgc:136866 | 0.64 |
| acp1 | 0.64 |
| zgc:158803 | 0.64 |
| thumpd1 | 0.63 |
| minal | 0.63 |
| ifrd2 | 0.63 |
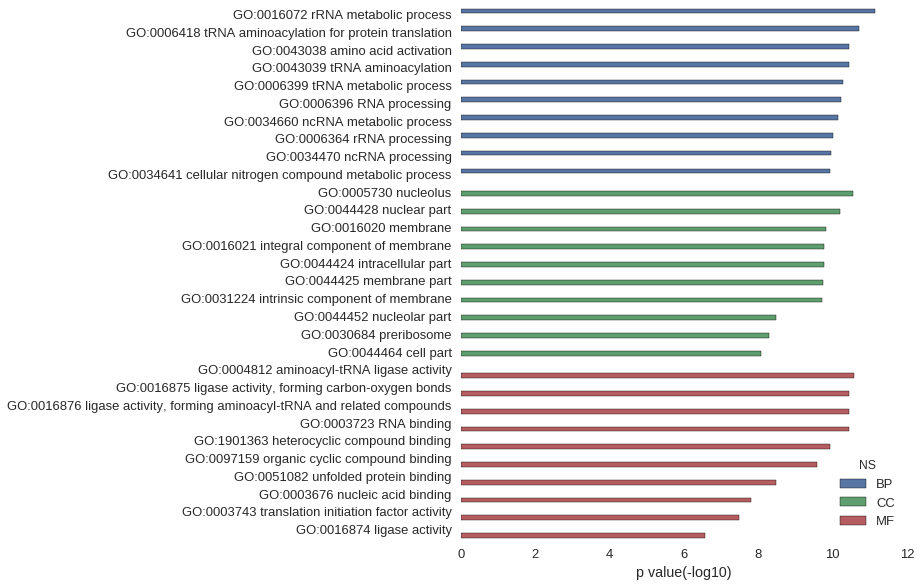
Gene Ontology
Download
| GO | P value |
|---|---|
| GO:0016072 | 7.29e-12 |
| GO:0006418 | 1.94e-11 |
| GO:0043038 | 3.56e-11 |
| GO:0043039 | 3.56e-11 |
| GO:0006399 | 5.20e-11 |
| GO:0006396 | 5.86e-11 |
| GO:0034660 | 7.16e-11 |
| GO:0006364 | 9.74e-11 |
| GO:0034470 | 1.08e-10 |
| GO:0034641 | 1.20e-10 |
| GO:0005730 | 2.80e-11 |
| GO:0044428 | 6.40e-11 |
| GO:0016020 | 1.55e-10 |
| GO:0016021 | 1.67e-10 |
| GO:0044424 | 1.73e-10 |
| GO:0044425 | 1.85e-10 |
| GO:0031224 | 1.94e-10 |
| GO:0044452 | 3.34e-09 |
| GO:0030684 | 5.16e-09 |
| GO:0044464 | 8.56e-09 |
| GO:0004812 | 2.64e-11 |
| GO:0016875 | 3.56e-11 |
| GO:0016876 | 3.56e-11 |
| GO:0003723 | 3.65e-11 |
| GO:1901363 | 1.20e-10 |
| GO:0097159 | 2.64e-10 |
| GO:0051082 | 3.34e-09 |
| GO:0003676 | 1.55e-08 |
| GO:0003743 | 3.31e-08 |
| GO:0016874 | 2.73e-07 |
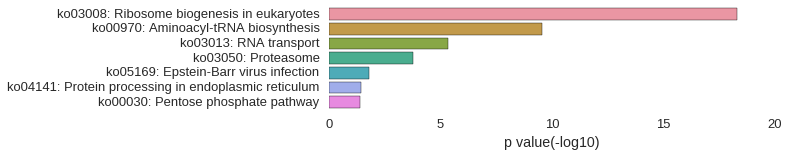
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03692
GAGAATATGCATCGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGTGTGCGCGTGCGTGCGTATGTGTG
TGTGTGTATGTTTATAAGAATAATATGGAGATCAGTGTGTGTTTGTCATCTCTGCGTCTGCATTAAAGGT
GTGTGTTGTTCAGCTGAAGCTCTGTTGACGCCAGTGGCAGAACATTATTACCACAGTAAATCATTTCCAG
CGCTGTTTTTAATAAATGCAGGAAGCATGAACATTGACTGCACTGATGGAAGTGAAACACACAACGTTAT
ACACACCTGGACACTGACGTGATGTAATGATCGTAATTCTGTCCAGTCATTGTCACCGTGACAGGTTCTG
CTGATAATGCTAGTGTTGCTGGCCAATATTGGAGAGTGTTGTGTCTTTATTATTGAGAAAGGCCAACAGA
TATTGTGTTTGTGTCCCATTGGCCTGGTTAGTTGCCCTGTGCCACACCTGGTTGCCCATTGATGACATCA
TTTGCCCAGCGGCATTTCTCAGCATTAGTCATGAGTTAATGTCTGATCATCGAGAGGTTTACTCCACCTG
AAGGACCCGGGGGAAATGGATTAAAGGAATTCATGTGTTAACCACTGGACTATTTGTGTTATTGCTCAAA
TCATAAACTAAATAAATGTAAAATAATAATAATTTTGCTCAAATAATAAATTGATTTGTC
GAGAATATGCATCGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGTGTGCGCGTGCGTGCGTATGTGTG
TGTGTGTATGTTTATAAGAATAATATGGAGATCAGTGTGTGTTTGTCATCTCTGCGTCTGCATTAAAGGT
GTGTGTTGTTCAGCTGAAGCTCTGTTGACGCCAGTGGCAGAACATTATTACCACAGTAAATCATTTCCAG
CGCTGTTTTTAATAAATGCAGGAAGCATGAACATTGACTGCACTGATGGAAGTGAAACACACAACGTTAT
ACACACCTGGACACTGACGTGATGTAATGATCGTAATTCTGTCCAGTCATTGTCACCGTGACAGGTTCTG
CTGATAATGCTAGTGTTGCTGGCCAATATTGGAGAGTGTTGTGTCTTTATTATTGAGAAAGGCCAACAGA
TATTGTGTTTGTGTCCCATTGGCCTGGTTAGTTGCCCTGTGCCACACCTGGTTGCCCATTGATGACATCA
TTTGCCCAGCGGCATTTCTCAGCATTAGTCATGAGTTAATGTCTGATCATCGAGAGGTTTACTCCACCTG
AAGGACCCGGGGGAAATGGATTAAAGGAATTCATGTGTTAACCACTGGACTATTTGTGTTATTGCTCAAA
TCATAAACTAAATAAATGTAAAATAATAATAATTTTGCTCAAATAATAAATTGATTTGTC